BioBye 029: solutions to RNA manufacturing, peptide-assisted genome editing, the biosignatures of dietary supplements, gene expression in the living human brain
Welcome to Decoding Bio, a writing collective focused on the latest scientific advancements, news, and people building at the intersection of tech x bio. If you’d like to connect or collaborate, please shoot us a note here or chat with us on Twitter: @ameekapadia @ketanyerneni @morgancheatham @pablolubroth @patricksmalone. Happy decoding!
It’s been an exciting week! A number of deals in the space: acquisitions by Recursion, partnerships with Ginkgo and successful IPOs. Let’s see what we have in store this week:
What we read
Blogs
Unlocking the Power of RNA: Overcoming Manufacturing Challenges [Nima Ronaghi, 2023]
Friend of the newsletter Nima Ronaghi published an essay this week detailing his firm’s investment thesis in EnPlusOne, an RNA enzymatic synthesis platform. RNA is an emerging therapeutic modality with a number of advantages, such as target specificity (small interfering RNAs or antisense oligonucleotides), rapid design, and streamlined manufacturing. RNA therapeutics also possess chemical and structural modularity, enabling modifications to improve their stability, potency, and delivery properties.
Chemical and enzymatic synthesis are the two primary methods for making RNA. Chemical methods have a number of important limitations including inefficient synthesis of RNA sequences, inability to make nucleic acid modifications for stabilization and delivery purposes, and limited scale of production (< 1 kg). EnPlusOne is pursuing an enzymatic strategy, which is more scalable than phosphoramidite synthesis. Spun out of the Wyss and incubated by Northpond Ventures, EnPlusOne has invented a de novo enzymatic process that produces RNA without requiring a DNA template. This expands the space of RNA that can be designed, for example by allowing the incorporation of modified nucleotides to extend the half-life of RNA. Further, EnPlusOne’s reactions are water-based, reducing the use of environmental harmful chemicals.
Biotech 2.0 [Neil Thanedar, 2023]
Neil Thanedar, a biotech investor, writes about this new era of biotech that we are entering (Biotech 2.0) by looking back on parallels in the internet and software age. He argues we need some tangible breakthroughs to make the leap from Biotech 1.0 to Biotech 2.0, in the same way that web 1.0 → web 2.0 was accelerated by:
Growth of new social networks
Creation of startup accelerators
Rise of institutional seed funds
Launch of first iPhone
In Neil’s words “if Biotech 1.0 was defined by static, centralized, mass-produced, one-size-fits-all products, Biotech 2.0 will be identified by dynamic code that uses each body as a platform to create its own solutions…and the technology is finally ready”. This article is part of his four part series on comparing the tech and biotech revolutions. He touches on the potential of what biotech 2.0 can unlock and opportunities in building and funding such companies.
Academic papers
Efficient engineering of human and mouse primary cells using peptide-assisted genome editing [Zhang et al., Nature Biotechnology, 2023]
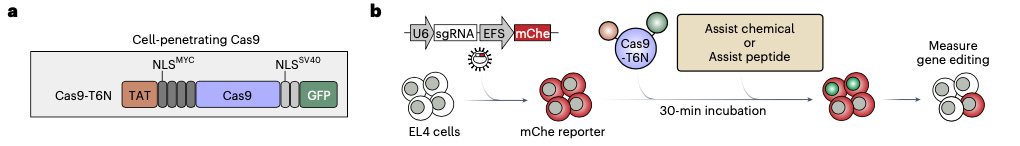
Why it matters: ex vivo delivery of CRISPR systems rely upon electroporation or viral vectors which cause cellular toxicity, transcriptional perturbation, long workflows and poor efficacy. The authors have developed a method to improve the uptake of Cas9 or Cas12a to significantly improve gene editing efficiency, whilst decreasing toxicity. This could improve the workflows of ex vivo engineered adoptive T cell therapies, cell reprogramming and in vivo genetic studies using mouse T cells in disease models.In addition to electroporation and viral delivery, several groups have attempted at cell-penetrating peptide (CPP) approaches that can potentially facilitate cellular uptake of the molecular cargo through endocytosis. Nonetheless, the cumulative inefficiencies of multiple cellular localisation events (i.e. cell entry, endosomal escape and nuclear localisation) have prohibited the wide adoption of these approaches in primary cells.
The authors of this study engineered a Peptide-Assisted Genome Editing (PAGE) CRISPR–Cas system for “facile, efficient, and nontoxic genome editing of primary cells”. The PAGE system consists of Cas9 or Cas12a and a CPP.
The group landed on the CPP protein fusion named TAT-HA2 that acts as an ‘assist peptide’. TAT-HA2 potentiated Cas9 editing of an mChe reporter line from an undetectable level to 85% efficiency and enabled gene editing at a tenfold lower concentration of the Cas9 protein with minimal loss of editing efficiency and minimal cellular toxicity. The system also demonstrated 80-90% gene editing efficiency in both natural killer human cell lines and primary human T cells, showing its ability to work with diverse cells.
The PAGE system was also applied to the development of CAR-Ts using Cas12a instead of the larger Cas9 protein. In a head-to-head comparison, they showed that the PAGE system showed 98% cell recovery versus 68% from electroporation treatment, thus showing lower cytotoxicity. Additionally, the opCas12a-RNP-PAGE-edited CAR-T cells showed no significant gene expression changes, in contrast with the thousands of gene expression changes in electroporated cells, indicating overall that PAGE is a better method to edit CAR-Ts.
Whilst the system still requires further optimisation and further evaluation of its immunogenicity, initial data suggest a strong contender for SOTA primary cell editing given its aforementioned advantages over electroporation and viral delivery.
A study of gene expression in the living human brain [Liharska, medRxiv, 2023]
Most molecular studies of the human brain have focused on post-mortem tissue samples rather than brain samples from living humans. This recent preprint from a group at Mt. Sinai tests the assumption that gene expression is unchanged in post-mortem brain tissue samples, and is therefore an appropriate proxy for in vivo biology.
Gene expression in the human prefrontal cortex was compared in 275 living samples and 243 postmortem samples. Samples were obtained from living donors during deep brain stimulation surgery for neurological diseases such as Parkinson’s disease. Gene expression levels differed for ~80% of the genes measured. Unsurprisingly, the largest differences in expression between living vs post-mortem samples was in genes involved in protein translation and cellular respiration and metabolism. More worryingly, postmortem brain gene expression signatures of Alzheimer’s disease, schizophrenia, Parkinson’s disease, and several other diseases were shown to be inaccurate representations of disease processes occurring in the living brain. Easier said than done, but going forward, human brain tissue (such as samples acquired during neurosurgery) should be prioritized for molecular studies of the human brain.
Glioblastoma remodelling of human neural circuits decreases survival [Krishna et al., Nature, 2023]
Why it matters: Glioblastoma remains the most common and aggressive brain cancer, with an abysmal prognosis for survival. Glioblastoma is known to be highly heterogeneous, complex, and particularly resistant to emerging therapeutics; indeed, the mechanisms underlying its lethality remain poorly understood. In this paper, Krishna et al. demonstrate that glioblastoma remodel neural circuits in the human brain, which promotes tumor progression, impaired cognition, and decreased survival in patients.The glial-neuron interface is complex; only recently has a significant body of work shown that gliomas interact with neurons, with bi-directional communication influencing tumor growth along with neuronal hyperexcitability amongst other mechanisms. In this paper, Krishna et al. used intraoperative electrocorticography (ECoG) while patients were engaged in language tasks (awake brain surgery) to understand the interplay between cortical and tumor dynamics. Interestingly enough, they found that tumor-infiltrated brain regions that were quite remote from known language regions functionally parasitizes neuroplasticity, without adding extra cognitive capabilities to distinguish between simple and complex words.
Additionally, the authors conducted a multi-modal analyses that demonstrated that tumors possessed high or low functional connectivity (HFC or LFC), in which HFC regions demonstrated higher expression of genes involved in neural circuit assembly, including TSP-1 while patients were engaged in language tasks (awake brain surgery).
A Draft Human Pangenome Reference [Liao et al., Nature, 2023]
Why it matters: creating a reference human genome should take into consideration genetic diversity that gives rise to the differences that make us unique. Pangenome studies present an opportunity to create a more comprehensive map of the human genome as it moves away from looking at one individual genome and instead combines data from many. Despite the fanfare of the human genome project reaching completion in 2003 and then again in 2022, we’re learning that mapping the human genome is just a constant work in progress. Researchers just announced a pangenome map that takes into account the complete DNA of 47 diverse individuals from across the globe into one genetic atlas. More and more humans will be added to this pangenome, with the argument being a single human genome does not take into account the sheer genetic diversity associated with the human race. Analyzing this set of data sheds light on evolutionary hot spots, gene deletions, and other artifacts that can help us redefine disease and find better ways to target them. Having a pangenome map makes gene research more equitable and provides reference on the parts of the genome that make us unique. This research was only possible with newer sequencing tools like Pacific Bioscience’s long read sequencing.
Quantitative Bioactivity Signatures of Dietary Supplements and Natural Products [Yasgar et al., ACS Pharmacology, 2023]
Why it matters: supplement culture is strong with dozens of new dietary compounds entering the market each week. Are these supplements useful? Do they actually compare to medicine? An ACS study investigates…We’ve all fallen prey to the advertising and hype behind certain nutritional supplements. Enter any grocery store or pharmacy (or even Instagram ads now) and you’re bombarded with various natural products from root extracts to tree bark all touting various health benefits and the fine print saying “These statements have not been evaluated by the FDA”. How many of these supplements are actually biologically active and functional? How many of these are borderline toxic? A recent study by the American Chemical Society quantified the bioactivity signatures of 300 dietary supplement and natural products (DSNP) and 664 traditional Chinese medicine (TCM) compounds. As expected, the results were mixed and pointed towards TCM compounds being more bioactive than DSNP. When run against liver metabolizing enzymes, 36% of DSNP compounds showed no activity with the others showing varying levels of selective and panel activity. To no surprise, many of the active compounds that come from a root or plant displayed activity when isolated but the root/plant itself showed no activity—showing that the actual product has different concentrations of the active ingredient which leads to differences in how that gets metabolized. Interestingly, the opposite is also true—mixes of plant components show activity but the individual components do not. Overall, the results of this study are varied and reinforce what we already knew—not all supplements are bioactive and effective (and they certainly don’t undergo close to the rigor of testing we reserve for drugs, despite advertising making us think otherwise).
Notable Deals
Recursion to acquire two Canadian drug discovery startups
In case you missed it
How will generative AI disrupt data science in drug discovery? [Jean-Philippe Vert, Nature Biotechnology Correspondence, 2023]
A Tale of Two Incretins [Dylan Neel, Kirti Nath, Harim Won, Biomarker, 2023]
What we liked on Twitter
Keeping $RXRX as my 4th place pick for #Techbio. Here is my rational.
Field Trip
Did we miss anything? Would you like to contribute to Decoding Bio by writing a guest post? Drop us a note here or chat with us on Twitter: @ameekapadia @ketanyerneni @morgancheatham @pablolubroth @patricksmalone