BioByte 016
LLMs for protein families, Nature's stance on GPT authors, deep learning for zinc finger design, T cell mania
Welcome to Decoding Bio, a writing collective focused on the latest scientific advancements, news, and people building at the intersection of tech x bio. If you’d like to connect or collaborate, please shoot us a note here. Happy decoding!
We’re officially 8% of the way through 2023. When did that happen?
After a whirlwind start to the year, we wanted to share a few updates on behalf of our team.
First, you may have noticed that we started calling ourselves “Decoding Bio” instead of “Decoding TechBio.” Why you might ask? We think the distinction (and derivative semantic arguments) are a distraction from our broader mission, which is to make information at the intersection of bioscience and computation more accessible. On the go-forward, we’ll be referring to ourselves as Decoding Bio and invite our community to help us embrace our new name.
Second, we took some time to synthesize our community’s feedback as we approach six months of writing this spring. Many of you shared that you enjoy the weekly round-up and “read it word for word” (wow). Others said they’d like us to experiment with long-form pieces on various topics.
We published our first long-form piece in early January with our 5 Bio Predictions, and again last week with our piece on Going Zero to One in TechBio. Over the next few months, we’ve got a number of exciting topical pieces in the hopper that we can’t wait to share. But, to distinguish between our long-form and short-form pieces, we’re going to refer to our weekly roundups as “BioBytes.” We get it, lots of ~re-branding~. We’re done for now.
Have any feedback on how we could make Decoding Bio better? Drop us a line at decodingbio@gmail.com.
Happy decoding!
What we read
Blogs
Tools such as ChatGPT threaten transparent science; here are our ground rules for their use [Nature, January 2023]
After a flurry of pre-prints surfaced listing ChatGPT as a co-author, Nature put a stake in the ground by launching ground rules for use of GPT in science. From Nature’s perspective, “The big worry in the research community is that students and scientists could deceitfully pass off LLM-written text as their own, or use LLMs in a simplistic fashion (such as to conduct an incomplete literature review) and produce work that is unreliable.” So here are the new rules:
No LLM tool will be accepted as a credited author on a research paper. That is because any attribution of authorship carries with it accountability for the work, and AI tools cannot take such responsibility.
Researchers using LLM tools should document this use in the methods or acknowledgments sections. If a paper does not include these sections, the introduction or another appropriate section can be used to document the use of the LLM.
The article also mentions work being done to detect the use of LLMs in text such as DetectGPT, which uses Zero-Shot Machine-Generated Text Detection using Probability Curvature.
What do you think? There have been some interesting debates on Twitter on this topic, and we expect other journals to respond.
Discovery moments: TYK2 pseudokinase inhibitor [Robert Pleng, 2023]
A fascinating essay chronicling the discovery of deucravacitinib, BMS' TYK2 inhibitor for psoriasis. The story is told in 3 parts:
Pivoting to a phenotypic screen to identify selective inhibitors. Rather than just focusing on the ATP-binding active site of TYK2, BMS developed a phenotypic screen that assessed the entire biological pathway of interest. This resulted in the unexpected discovery of a second TYK2 binding pocket, the pseudokinase (JH2) domain.
Elucidating the mechanism of TYK2 inhibition via modulation of the pseudokinase domain. Through a series of painstaking experiments, it was determined that the pseudokinase domain allosterically regulated the TYK2 catalytic site.
The ol’ lead op switcheroo. The BMS team identified a novel hit for the TYK2 pseudokinase domain but discovered when the molecule was tested in vivo that a less selective metabolite was produced. To solve this problem, scientists performed a deuterium switcheroo (changing 3 H atoms for 3 deuteriums), which reduced the undesirable metabolic pathway.
Engineering T Cells [Ground Truths, Eric Topol, 2023]
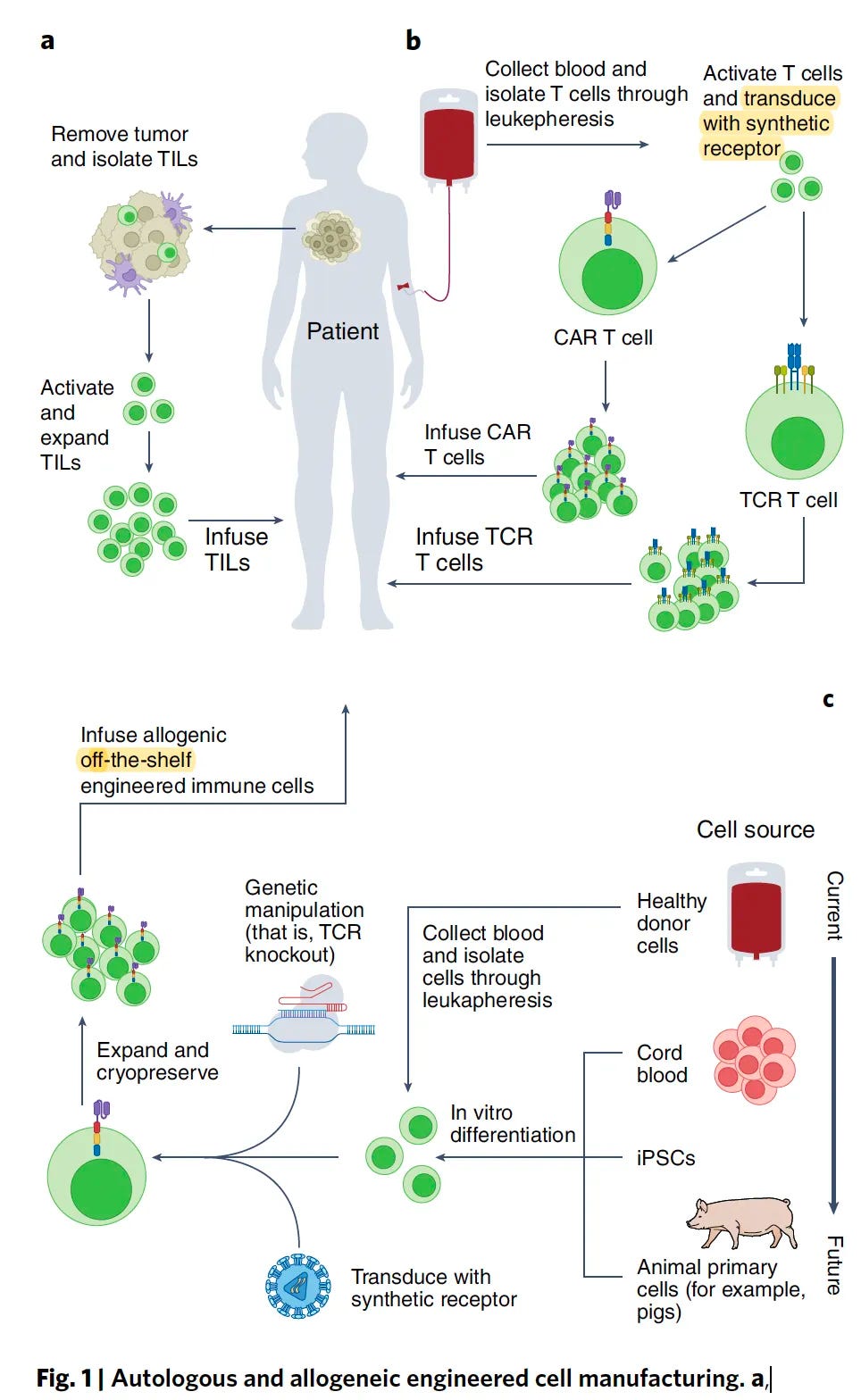
In his recent issue of Ground Truths, Dr. Eric Topol provides an overview of the various ways T cells are being used to treat disease: via both up and downregulation of the immune system across various cancers, autoimmune diseases, and even multiple sclerosis. As a refresher, CAR-T cell engineering is a promising form of personalized medicine that modifies T cells to recognize proteins in a person's cells and in effect, modulates the resultant immune response. As Dr. Topol highlights, there are over 500 clinical trials ongoing, building on the first CAR-T cell treatment approved for leukemia just six years ago.
Despite immense progress in T-cell engineering, the piece rightly highlights how costs remain prohibitive for CAR-T to become a mainstream therapy: “approximately $500,000 for a single treatment, which doesn’t include the patient’s hospitalization and other required treatments.” The piece concludes with a mention of an emerging area of work, notably “off the shelf CAR-T” (vs. autologous).
A Catalog of Big Visions for Biology [Sam Rodriques]
Sam’s latest post admittedly poses more questions than answers but the open-ended type that makes you think a little bit harder about the world and question how we do things today. Sam’s thoughts are predicated by one claim-that grand visions drive humanity to do great things. In biology, grand visions often come with heavy contextualization and caveats which are justifiable given the constraints of biology and clinical process. But what if we dreamed beyond what we currently know? We found reading through Sam’s open-ended questions for biological progress inspiring.
Academic papers
Large language models generate functional protein sequences across diverse families [Nature Biotechnology, January 2023]
Why it matters: LLMs can learn to generate protein sequences with a predictable function across large protein families. These models have demonstrated artificial enzymes from scratch, which, in laboratory tests, appear to work as well as those found in nature (even despite divergent amino acid sequences not found in nature). This technology developed in this paper has spawned a new company called Profluent Bio founded by Ali Madani PhD.Big splash in large language for protein models this week! Researchers have developed ProGen, a deep-learning language model that can generate protein sequences with predictable functions across large protein families, similar to generating grammatically and semantically correct natural language sentences on diverse topics. The model was trained on 280 million protein sequences from over 19,000 families and is controlled by tags specifying protein properties. ProGen can be fine-tuned to improve its performance for specific protein families and has been demonstrated to generate artificial proteins with similar efficiencies to natural proteins.
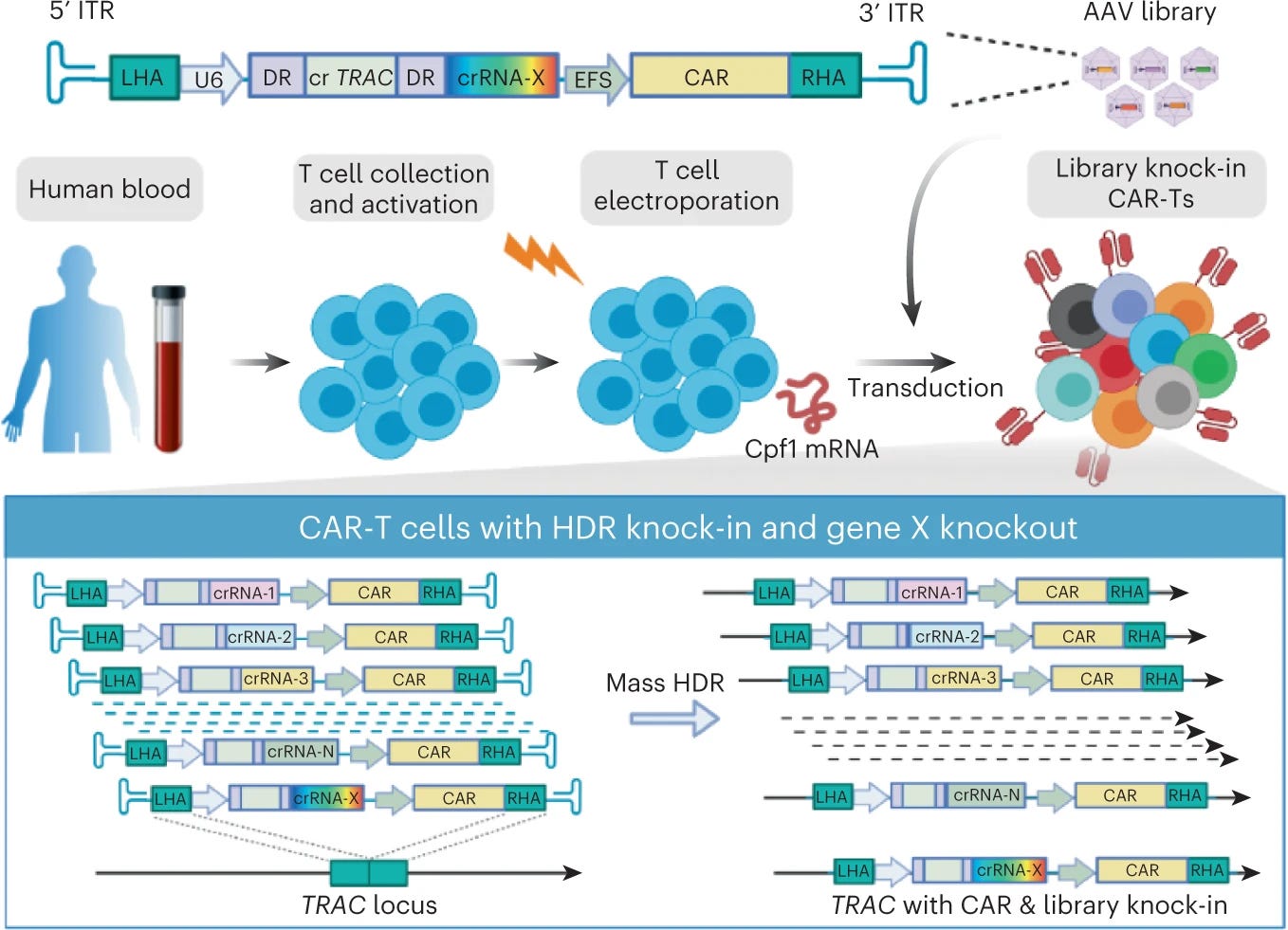
Massively parallel knock-in engineering of human T-cells [Dai et al., Nature Biotechnology, 2022]
Why it matters: CLASH can enable the generation of a sizable number of genomic knock-ins in parallel, representing a step change in cell engineering throughput. With adaptations, CLASH may be applied to any other cell type, breaching the limits of what was capable for high throughput engineering, and enabling faster design-build-test-learn cycles. For years, approaches to engineering cell therapies have been low throughput, lacking the ability to introduce and assess a combinatorial suite of edits. CRISPR-based T-cell screens use viral vectors or transposons to integrate DNA sequences of choice; these can lead to insertional mutagenesis, downstream translational silencing, and lower-than-ideal efficiencies.
In this paper, Dai et al. develop CLASH – CRISPR-based library-scale AAV perturbation with simultaneous HDR knock-in, to engineer T-cells. Briefly, they delivered mRNA encoding Cas12a (enzyme for gene editing) via electroporation and used AAV to deliver the Cas12a CRISPR RNA array + knock-in transgene cargoes. Splitting delivery serves a major purpose here: the Cas12a mRNA is always constant, while AAV vector libraries can be designed and scaled for synthesis, enabling multiplexed edits of choice. The crRNAs and transgenes integrate into a parallel fashion into the TRAC locus by AAV-mediated HDR. Their initial proof of concept work generated large pools of CAR-T cell variants simultaneously, which may unlock a number of therapeutically-relevant constructs.
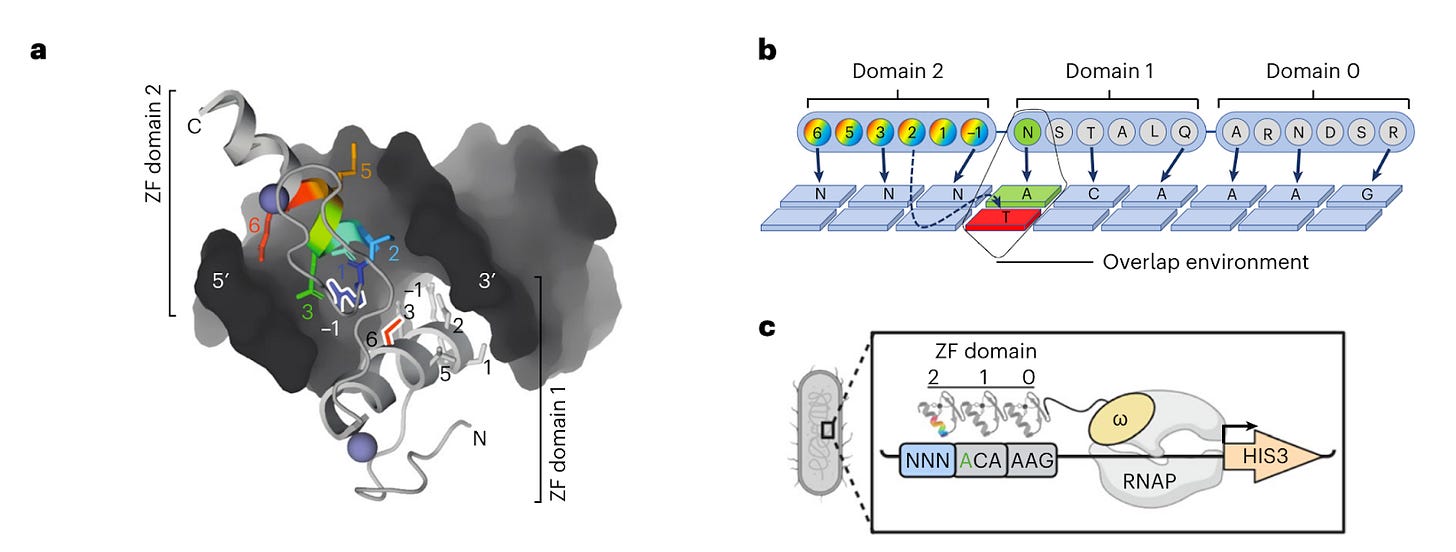
A universal deep-learning model for zinc finger design enables transcription factor reprogramming [Ichikawa et al., Nature Biotechnology, 2023]
The authors, from NYU and University of Toronto, have developed ZFDesign, a deep learning model that can design zinc finger domains to attach to any section of DNA, inducing either activation or repression of a specific gene.
The model was trained by screening 49 billion protein-DNA interactions, with the aim to utilize the platform to identify ZF domains that can modulate gene expression in order to treat diseases caused by haploinsufficiency or gain-of-function mutations.
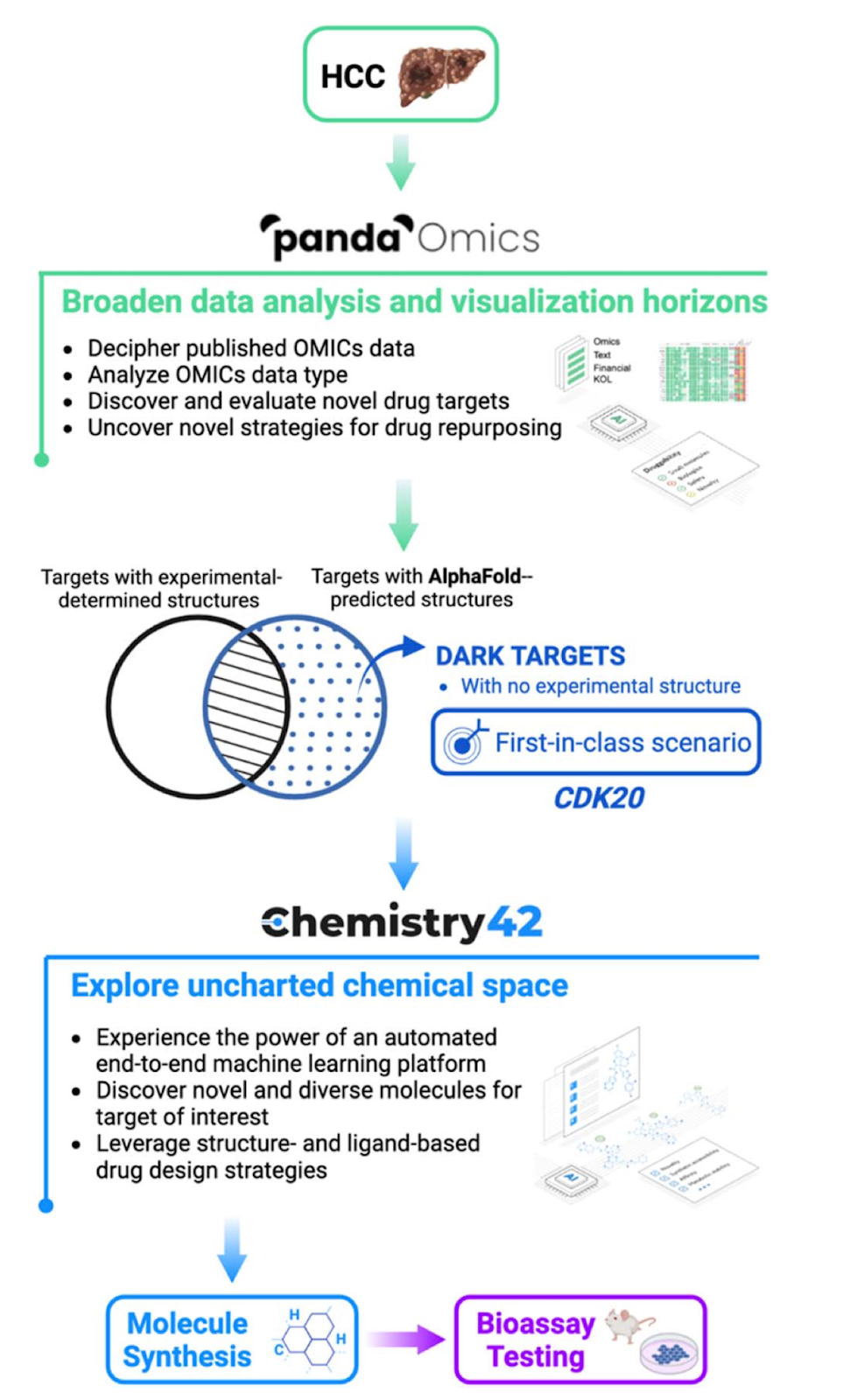
AlphaFold accelerates artificial intelligence powered drug discovery: efficient discovery of a novel CDK20 small molecule inhibitor [Ren et al., Chemical Science, 2022]
Insilico Medicine published the first paper (as far as we’re aware) describing the discovery of a hit for a novel target using AlphaFold without the use of an experimentally derived structure. A new target for hepatocellular carcinoma (cyclin-dependent kinase 20) was identified using Insilico’s multi-omics database, and structure-based generative chemistry was used to generate candidate hits. It took only 30 days and the synthesis of 7 compounds to discover a suitable hit. Importantly, lead optimization or ADME testing was not performed but will be important next steps.
What we listened to
Notable Deals
Profluent debuts to design proteins with machine learning in bid to move past 'AI sprinkled on top'
Three VCs launch Dimension, a new firm with plans to fuel biotech’s ‘digitization’
Colossal Biosciences Secures $150M Series B and Announces Plan to De-Extinct the Iconic Dodo
In case you missed it
Britain is losing its chance to become a life sciences superpower
Federated machine learning in data-protection-compliant research
What we liked on Twitter









Field Trip
Did we miss anything? Would you like to contribute to Decoding Bio by writing a guest post? Drop us a note here or chat with us on Twitter: @ameekapadia @ketanyerneni @morgancheatham @pablolubroth @patricksmalone














I like the name change!