BioByte 021: the future of drug blockbusters, tiktok and protein design, crowdsourcing RNA degradation predictions, gripping integrins
Welcome to Decoding Bio, a writing collective focused on the latest scientific advancements, news, and people building at the intersection of tech x bio. If you’d like to connect or collaborate, please shoot us a note here. Happy decoding!
Welcome back to BioByte, your one-stop-shop to find peace, joy and biotech. Let’s dive in.
What we read
Blogs
Pharmaceutical blockbusters: the past, present, and future [Alex Telford, 2023]
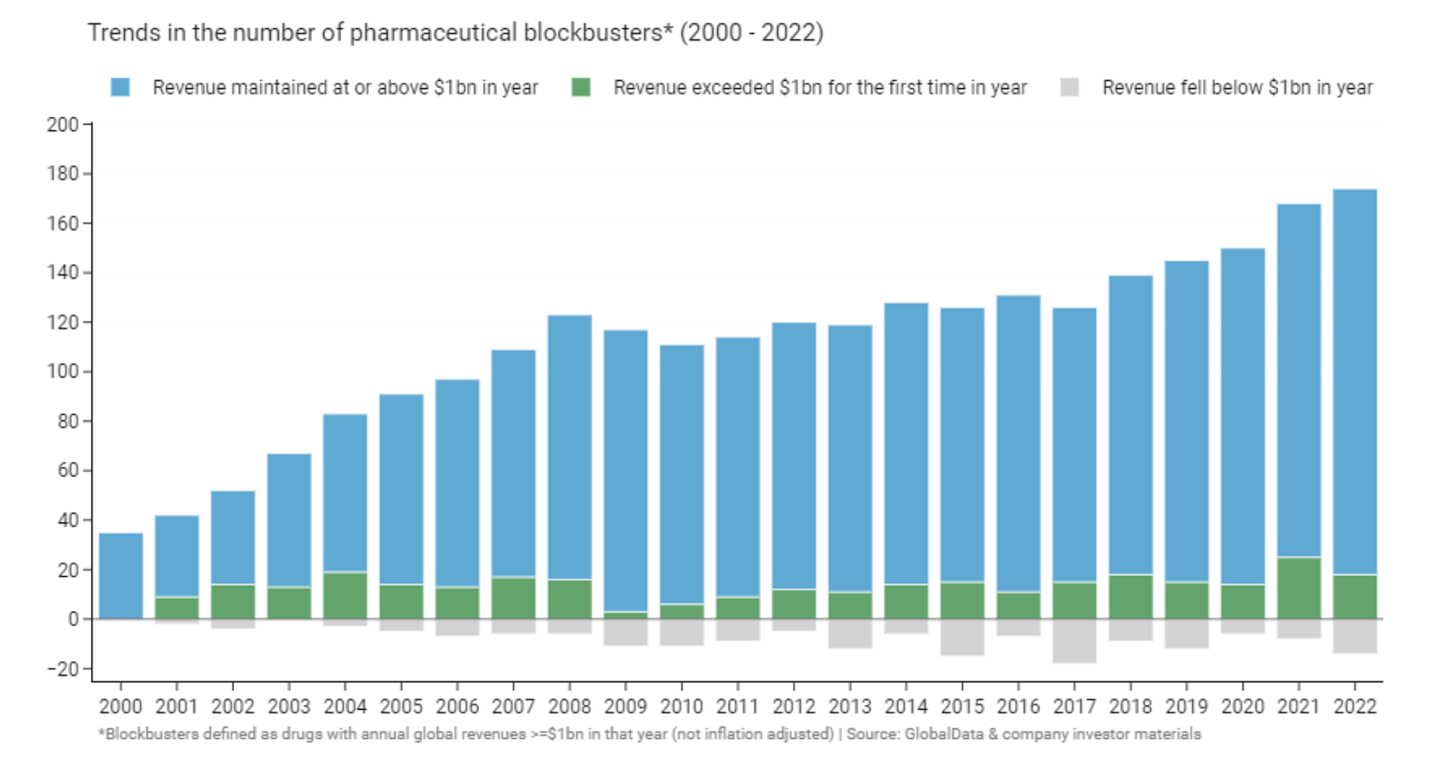
Given the high failure rate of drug development, biopharma is heavily reliant on blockbusters (drugs with >$1 billion in revenue). The number of blockbuster drugs, and the total proportion of pharma revenue resulting from blockbusters, has increased over time (figure below). Only 21% of drugs approved between 2011 and 2022 by the 20 largest pharma were blockbusters, yet they accounted for ~70% of revenues for these companies.
What are the common characteristics across blockbuster drugs? Drugs that are fast-followers that come to market 2-5 years after a first-in-class drug tend to be the biggest sellers and are more likely to be blockbusters. It pays to be best-in-class, not first-in-class. This partially helps explain why positive clinical data that de-risks novel biology in a historically graveyard indication or modality results in a flurry of investment and activity as others rush to follow (I have a weird mental model that likens drug development to a minefield).

Alex documents several other trends:
Despite the 20 year patent life of Humira, in general the average age of blockbusters has not changed much since 2000.
The number of biologic blockbuster drugs has steadily grown and now rivals the number of small molecules.
The number of rare disease drugs has increased to the point that 57% of drugs approved by the FDA in 2022 were orphan drugs. This trend was likely due to the US Orphan Drug Act and the rise of genetic medicine capable of precisely targeting genetically-driven rare disease.
Since 2000, blockbusters have gradually shifted from primary (e.g., hyperlipidemia) to speciality (autoimmune, oncology).
Now for the interesting part: Alex’s predictions for the future, as well as some commentary (see the full article for all of the arguments/evidence for each prediction).
The fundamental economics of the industry stay much the same, and drug revenues continue to be driven primarily by blockbusters.
In the near/medium term, I agree that financial incentives will encourage the development of more blockbusters. But over the long term, I wonder if precision medicine and personalized targeting of drugs to specific (and smaller) patient sub-populations will gradually reduce the TAM for each individual drug, resulting in a fragmented competitive landscape where some drugs work for some patients some of the time. Prediction #5 below instead imagines that personalized medicine drives blockbuster status to the platform (which produces personalized therapeutic products for each patient) rather than the drug itself.
It gets harder to defend blockbuster revenues.
There will be a stronger focus on commercial/clinical defensibility going forward such as companion diagnostics, branding and marketing, strategic partnerships with payers, providers, or patient groups, etc.
Out of favor indications make a comeback.
For the sake of our future, let’s hope antibiotics become fashionable sooner rather than later.
Orphan drug prices come under pressure, and some orphan blockbusters fall off.
Blockbuster drugs get more diverse and personalized, platforms become blockbuster products.
Facebook, TikTok, and proteins: Why big tech companies are betting on AI protein design [Brittany Trang, STAT News, 2023]
We’ve previously discussed the work coming out of the research arms of big tech companies and mentioned it as one of the major themes in 2023. But why are these tech giants interested in the life sciences? Why not devote all those resources, especially given the cost-cutting, to the core aspects of their business?
This STAT News article shares some diverse opinions:
“It’s, in a sense, a low-hanging fruit. It’s kind of an obvious problem for the tools that now exist,” said Simona Cristea, the head of data science at Dana-Farber Cancer Institute’s pancreatic cancer research center. “It’s easy to formulate the problem…whereas with other questions in biology, you cannot formulate the problem. These are not problems that you just apply a deep learning model to.”
“It’s part of our broader agenda of giving back to society using technology that we’re building anyway for Salesforce applications, but can be applied to problems for societal impact as well,” said Nikhil Naik, director of research at Salesforce and an author on the Nature Biotech paper.
“AI is really critical to the future of the company. It’s something that Mark Zuckerberg and other leaders of the company have talked about,” said Rives. While the team is exploring language models that might have applications across AI domains, biology offers the chance to test how well the AI can learn on a very complex level. Whereas humans can check language models by simply deciding whether a sentence sounds like a sentence or not, protein structure and function depend on a much more complex set of variables that scientists themselves don’t yet fully understand.
We don’t take product goals; our goals are to advance the science,” said Rives at Meta
The impact of the protein structure and function research coming out of these research labs is invaluable to the field. Whilst the impact is global, there are obvious advantages for the firms as their learnings can also be adapted to other, more commercialisable products.
Patients are the Platform [Perlara PBC, 2023]
We’ve talked a lot about how biology is slowed down by inefficient clinical trials, and at the root of that problem often lies patient recruitment. Ethan Perlstein argues that “patients are the platform” in this new piece. He claims that patient communities drive the formation of new medicine—from advocacy to clinical trial participation to being the group that makes up the end users. So with this in mind, the big-pharma and investor-backed companies that spend millions of dollars before committing to an indication is unsustainable. With platforms becoming increasingly commoditized as technology advances, Ethan argues that the pace of innovation is no longer the bottleneck, the “ultimate rate-limiting step for any new medicine will always be a pivotal Phase 3 study approval, which is in the hands of 30-40 volunteer study participants, if you’re the average rare disease”. As such, patients become the discovery platform (disease models, drug screens, biomarkers, etc), the clinical platform (studies), and the commercial platforms (communities, trials). As we think about how to unlock efficiencies in biopharma, we suspect this line of thinking will gain traction.
Academic papers
Crowdsourcing to predict RNA degradation and secondary structure [Hendrix, Nature Machine Intelligence, 2023]
Why it matters: Predicting RNA degradation is fundamental to design stable RNA-based therapeutics. In the paper, the authors use crowdsourcing is a way of gathering many hypotheses and models to find the best predictors of RNA degradation. These models could be useful in cases where one needs to predict degradation kinetics such as circadian gene expression research. Wayment-Steel et al. combined two separate crowdsourcing projects to predict chemical stability of RNA based on sequence and structure information.
The first project was done via the Eterna website. Participants in the project designed RNA sequences to be highly stable against degradation. This crowdsourcing effort led to multiple hypotheses each with thousands of associated RNA sequences rather than a single group of scientists working on a single hypothesis.
The second crowdfunding project, organized via Kaggle, entailed users to develop machine learning models to predict RNA degradation patterns from the provided sequence and predicted structure data resulting from the first project. The two best performing models in Kaggle had a lot in common in that they both used layers of recurrent neural networks, using the RNA sequence and structure as input.
This model could be useful in cases where one needs to predict degradation kinetics. For instance in circadian gene expression research, where between the sample is taken post-mortem and when it is analyzed, there has been some degradation in RNA. This model would allow researchers to ‘turn back the clock’ and predict what was the original expression level.
The Gripping Story of Integrins [James Rothman, Cell, 2022]
A historical piece published in Cell last year on the history of integrins, a special class of proteins that span the cell membrane and are capable of both extracellular and intracellular communication. Integrins are heterodimers consisting of an alpha and beta subunit to create 24 unique integrin subtypes. Integrins facilitate cell-cell and cell-extracellular matrix adhesion, are involved in a wide range of biological activities and diseases, and since 2015, at least 130 integrin inhibitors have entered clinical trials, including several FDA-approved therapies.
The essay recounts three parallel research programs that eventually led to a comprehensive description of the integrin class of receptors. In 1977, Timothy Springer discovered that cytotoxic T cells detected their targets via integrin signaling, which we understand today to be combined antigen and cell recognition via antigen and integrin co-receptors. Separately, Richard Hynes discovered that the glycoprotein fibronectin was involved in behavior of cancerous cells, and acted through integrin receptors. Finally, Erkk Ruoslahti discovered that fibronectin bound to its receptor via a four amino acid domain, arginine-glycine-aspartate-serine (RGDS), and hypothesized that a larger family of RGD-binding cell-matrix receptors may exist. As Hyne recounts, the 3 scientists’ work culminated at a conference in 1987:
“It was another of those special moments in science. As it happened, I was organizing a Gordon Conference…for February 1987… It became clear at the Gordon Conference that there was indeed a family of receptors present in mammals, birds and insects…people from widely divergent fields met each other for the first time. Each field was approaching these receptors from different viewpoints…All of the sudden, each discipline had a whole new set of ideas.”
Hynes published an overview of integrins in 1987, which launched the field of integrin research.
Network expansion of genetic associations defines a pleiotropy map of human cell biology [Barrio-Hernandez et al., Nature Genetics, February 2023]
Why it matters: Protein interaction networks have long been known to be fertile ground for understanding gene associations. This paper demonstrated that network-based genome-wide association study approaches can help uncover genes and cellular processes linked to human traits. Analyzing pleotropic dynamics -- whereby a single gene or variant can impact multiple traits -- via network-based methods offers greater insight into biological pathways at the level of the individual organism, and also highlights new opportunities for drug development and repurposing.Leveraging >1,000 trait-associated genes, researchers identified groups of traits that are likely to share the underlying genetic and biological process. Leveraging a personalized PageRank algorithm that expanded genes likely to be causal for a given trait, communities of densely connected proteins were then associated via random walk clustering. As demonstrated by other studies, network expansion methods were successful in identifying previously known disease genes not identified by GWAS (including those not in GWAS loci but that may modulate the same biological processes).
The team then applied these approaches to inflammatory bowel disease to uncover inflammatory bowel (IBD) disease-relevant genes with strong functional and genetic support. These pathogenic variants were then mapped to targets of approved drugs to explore potential opportunities for repurposing.
What we listened to
Notable Deals
TandemAI raises $35M in Series A for small molecule discovery, wet labs
What we liked on Twitter





Events
Science2Startup | May 3rd 2023 | Boston
S2S is a forum for top scientists from around the world to present their ideas and interact with leading investors and executives in the Boston biotechnology hub.
Over 8 years, investigators from institutions including Brown, Carnegie Mellon, Columbia, Harvard, Johns Hopkins, MIT, Northwestern, NYU, Purdue, Scripps, UCLA, University of Iceland, University of Ottawa, University of Pennsylvania, University of Pittsburgh, Yale, and many more, have presented their startup ideas at S2S and its precursor, the University Research and Entrepreneurship Symposium (URES).
Terasaki Innovation Summit 2023 | March 8-10 2023 | LA
The mission of the Institute is to invent and foster practical solutions that restore or enhance individuals' health. We envision a world where personalized medicine is available to all. Innovation and quick translation of technology are a cornerstone of our mission and vision. The Inaugural Terasaki Innovation Summit will include key leading investigators discussing the latest advances in micro- and nanotechnologies and applications in diagnostics, therapeutic drug delivery, and regenerative medicine. In addition, key business leaders and experts in translation, innovation, IP, and more will be on hand to share their knowledge and commercial experience.
Field Trip
Did we miss anything? Would you like to contribute to Decoding Bio by writing a guest post? Drop us a note here or chat with us on Twitter: @ameekapadia @ketanyerneni @morgancheatham @pablolubroth @patricksmalone