BioByte 024: diffusion docking models, super evolution, organoid intelligence, frontiers in bio automation and cats
Welcome to Decoding Bio, a writing collective focused on the latest scientific advancements, news, and people building at the intersection of tech x bio. If you’d like to connect or collaborate, please shoot us a note here or chat with us on Twitter: @ameekapadia @ketanyerneni @morgancheatham @pablolubroth @patricksmalone. Happy decoding!

Hey, hey. This was a big week in bio, so let’s get to it. Only thing to note is that we will dropping a huge report soon. Keep your eyes peeled and emails subscribed.
What we read
Blogs
Speeding up drug discovery with diffusion generative models [Alex Ouyang, MIT News, 2023]
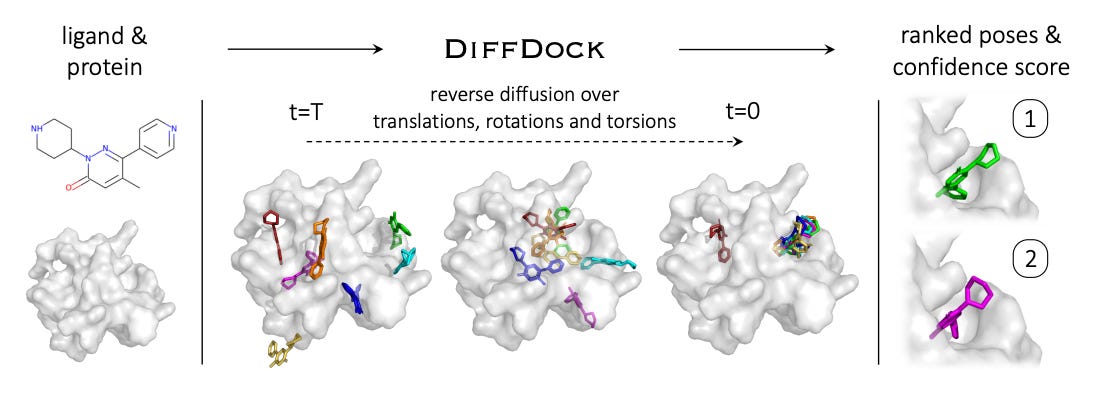
Most molecular docking tools used for in-silico drug design utilize a “sampling and scoring” approach, searching for the best fit “pose” of the ligand when bound to a target protein pocket. This is time-consuming due to the large number of configurations that have to be tested. Most deep-learning approaches to molecular docking treat it as a regression problem, implying there is a single right answer.
Gabriele Corso, co-author and MIT PhD student, states that “With generative modeling, you assume that there is a distribution of possible answers — this is critical in the presence of uncertainty.”
DiffDock allows multiple poses to be predicted with associated probabilities, rather than arriving at a single answer. The diffusion model DiffDock was trained on a variety of ligand and protein conformations, which enabled them to identify binding sites on proteins that it had never encountered before. DiffDock seems to be doubly as accurate as other docking models (22% vs 10% predictions within 2 angstroms when using computationally predicted structures).
Super evolution x Biology - 2023 Edition [Innovation Endeavors, 2023]
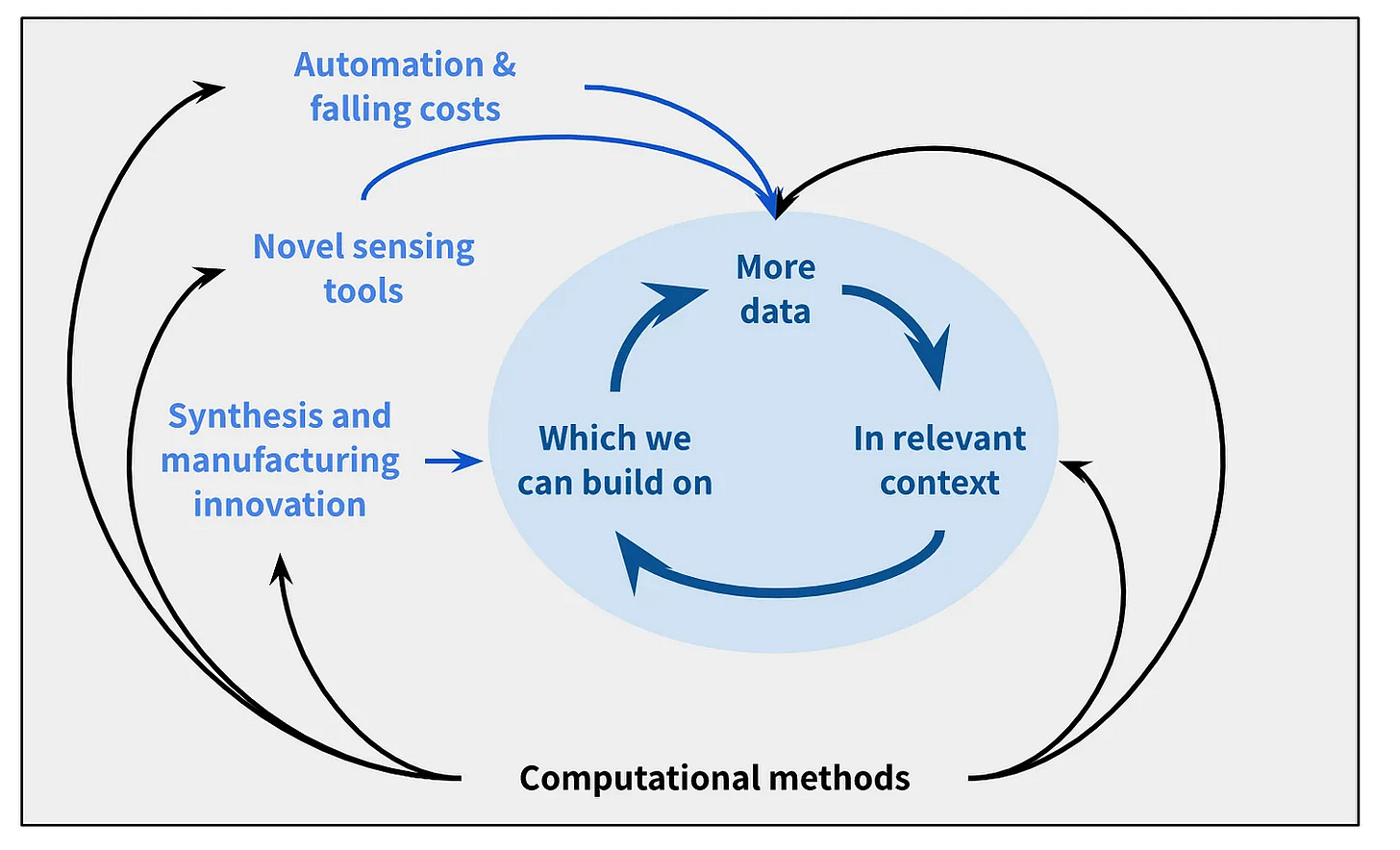
Nick Olsen, Carrie von Muench, and Galen Xing at Innovation Endeavors, an anti-reductionist, pro-complexity venture firm with a number of sectors of focus including bio, released an updated investment thesis. The common denominator across all of the companies IE gets excited about are technologies that combine computational methods with the ability to generate differentiated data in a biologically relevant context.
Differentiated data arises from 1) scaled data (data collection/generation with increased throughput using methods like droplet microfluidics) and/or 2) novel data (e.g., super-resolution microscopy to visualize protein motion). Additionally, data collected in its native biological context, or that directly characterizes function (rather than using a proxy) is most desirable.
Finally, a few spaces IE is excited to invest in:
Novel target biology identification and validation, especially targets with strong genetic underpinnings, or validated using preclinical models with strong predictive validity
Clinical translation tools (RWD, synthetic control arms, adaptive trials)
Next-generation therapeutic effects, such as new tools for engineering polypharmacological activity
Frontiers of Bio Automation [Shelby Newsad, Vega Shah]
Shelby and Vega reflect on SLAS, the annual convention for lab automation in this post. Fully-automated labs directed asynchronously have been promised for several years now and technology is converging to make this a conceivable reality. That would open up geographies (doing science in space and inhabitable climates) and lower the cost barriers to conducting experiments given a lot of the expenditure is related to lab management including things like hazardous waste disposal and procurement. They also noticed more companies attacking the immediate steps to lab automation that involve standardizing procedures like liquid handling, colony picking, and running basic experiments as well as the layer of software that enables this transition to take place. To conclude, they touch on a rather new concept of “citizen science” where human biohackers are essentially treated as individual experiment-generators, or labs. This concept takes decentralized science to the extreme and we’re intrigued.
Academic papers
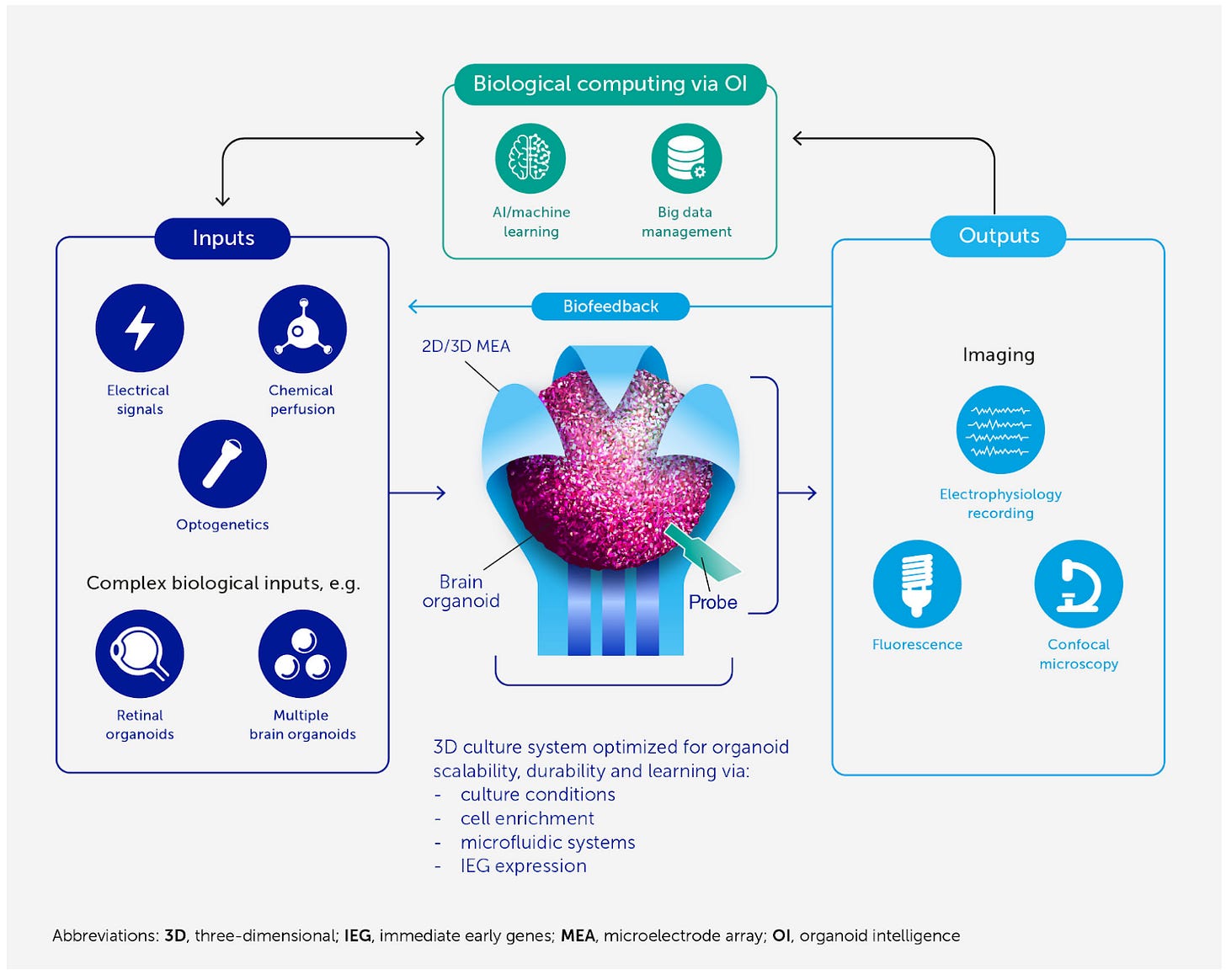
Organoid intelligence (OI): the new frontier in biocomputing and intelligence-in-a-dish [Smirnova et al., Frontiers in Science, 2023]
Yeah AI is cool, but have you heard of OI?
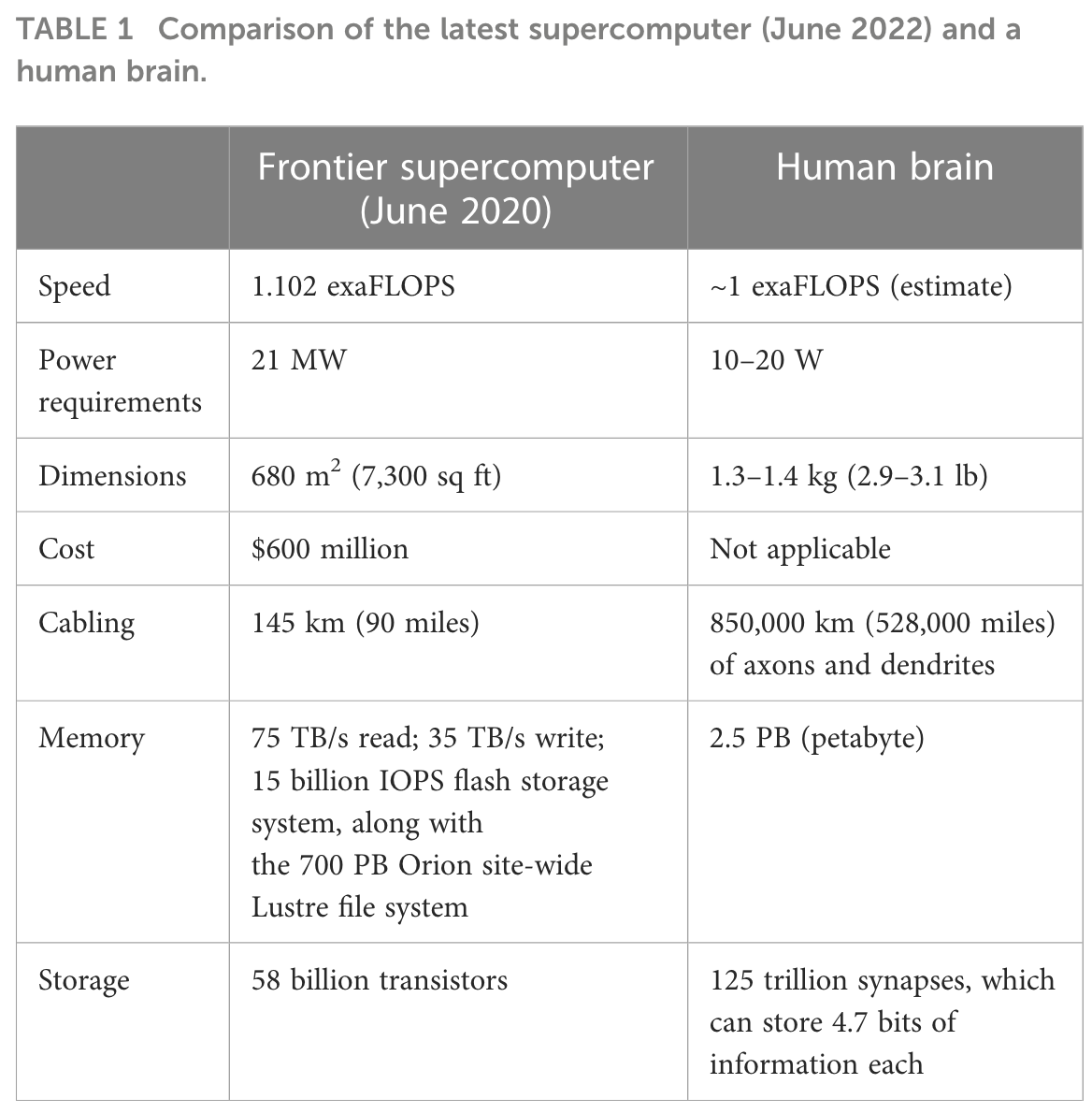
This new term encapsulates biological computing that harnesses brain organoids and brain-computer interfaces (BCIs). Human brains have two important efficiency advantages compared to computers: 1) biological learning uses far less power to solve computational problems and 2) it uses fewer observations to learn to solve problems. OI, 3D neuronal cultures paired with BCIs, aims to combine the advantage of computers to process information constantly with the efficiency advantages of biological computing. This symbiosis could enable us to unlock neuromimetic AI algorithms and develop better BCIs to treat neurodegenerative diseases.
Characterizing the interaction conformation between T-cell receptors and epitopes with deep learning [Nature Machine Intelligence, Peng et al., 2023]
Why it matters: Understanding the immune system holds the key to unlocking treatments to several classes of diseases including autoimmune, cancers, and infectious diseases. T cells mediate the immune response and are often targeted in cell therapies (such as CAR-T therapy) but have incredibly complex, poorly understood biology so understanding their binding and function is crucial to designing safer and more efficacious therapies.T-cell <> epitope recognition is a key part of the adaptive immune response, and the diversity of interaction between T cell and epitope (part of antigen) is vast. There are many projects underway to sequence T-cell and antigen interactions to be able to predict what T-cell receptors (TCRs) bind to which antigens and vice versa. This study pioneers a new deep learning framework to model the TCR-epitope interactions at a residue (spatial) level (TEIM-Res), which in practice looks at sequences of a bound TCR-epitope complex and in turn predicts the distances and contact probabilities of residues between part of the TCR and the epitope, taking sequence data and deriving structural context. They used a few-shot learning approach which means training their models on very little data given the lack of availability. With fine-tuning, this paper presents an approach to elucidating the interaction and structure of more TCR/antigen pairs, which can be used to design better vaccines and therapeutics that involve T-cell activation. It’s also possible to take the patterns this model may undercover and see if there are specific rules that govern TCR/epitope binding. Note: this work only encompasses CD8+ TCRs and MHC-I complexes, which are the T-cells involved in directly killing pathogenic cells. The scope should be expanded to CD4+/MHCII complexes in the future.
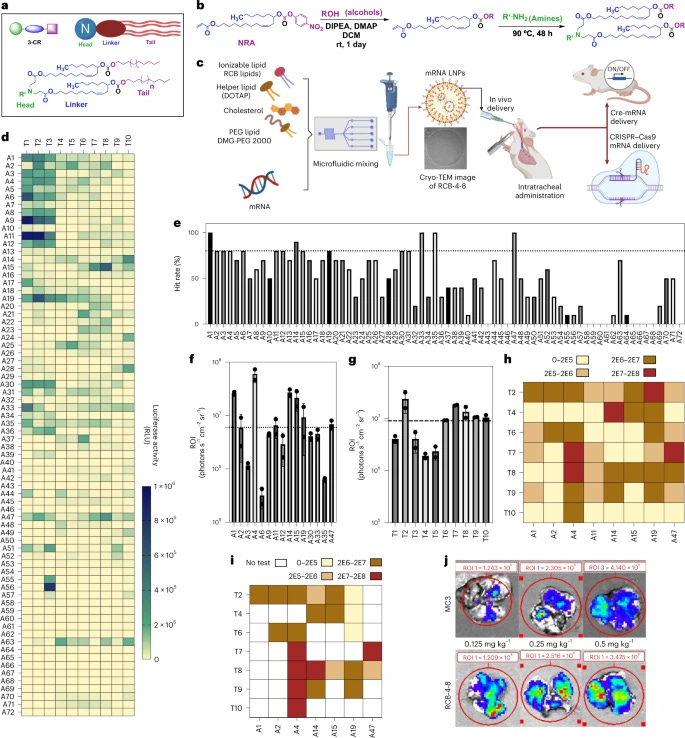
Combinatorial design of nanoparticles for pulmonary mRNA delivery and genome editing [Li et al., Nature Biotechnology, 2023]
Why it matters: Delivery of genetic medicines remains one of the most pressing challenges in medicine. Although viral vectors have demonstrated some degree of success, they are limited by immunogenicity concerns, poor tropism, and packaging sizes. In recent years, nonviral approaches such lipid nanoparticles (LNPs) nonviral approaches, such as lipid nanoparticles (LNPs) have garnered interest as they may overcome said challenges of viral vectors. The lung has remained a challenging target for gene therapy given the heterogeneity of cell types, mucous barrier, and lung clearance mechanisms. In this paper, Li et al develop lung-optimized LNPs to deliver gene editing modalities.
Briefly, the authors synthesized a library of ionizable lipids using a three-component reaction (3-CR) system. Here, a nitro ricinoleic acrylate (NRA) linker was coupled with aliphatic alcohols (lipids), which were connected to headgroups that were primary-tertiary amines. This system allowed for simplified generation of a combinatorial library, as compared to the previous 2-CR system (amines conjugated directly to lipids). They generated 720 new lipids with a variety of tails and headgroups and evaluated the potential of the LNPs to co-deliver cas9 mRNA and sgRNA for pulmonary gene editing. Remarkably, they found their lead LNP to improve mRNA transfection in mouse lung 100-fold greater than LNPs containing MC3, a lipid approved by the USFDA for RNA delivery. Additionally, they achieved editing in ciliated and club cells (both of which are implicated in congenital disease). This work is exciting as it introduces a powerful new tool in the toolkit, opening the doors for treating inherited lung diseases.
What we listened to
Notable Deals
Ginkgo partners up with Sensible Biotechnologies to forge mRNA manufacturing platform
Mosaic Therapeutics closes $28m series A funding and appoints Brian Gladsden as CEO
Mitochondria biotech field grows as RA, Insight up the Capacity with a new $35M startup
In case you missed it


What we liked on Twitter









Field Trip
Did we miss anything? Would you like to contribute to Decoding Bio by writing a guest post? Drop us a note here or chat with us on Twitter: @ameekapadia @ketanyerneni @morgancheatham @pablolubroth @patricksmalone