BioByte 030: legislation for gene-edited crops, bio-developer survey, foundation model for single cell biology, first non-hormonal menopause drug
Welcome to Decoding Bio, a writing collective focused on the latest scientific advancements, news, and people building at the intersection of tech x bio. If you’d like to connect or collaborate, please shoot us a note here or chat with us on Twitter: @ameekapadia @ketanyerneni @morgancheatham @pablolubroth @patricksmalone. Happy decoding!
Packed newsletter this week! If you only have a moment, here are the highlights:
New legislation in the UK that enables genetic technologies for agriculture and food production, permitting precision breeding using gene editing to engineer disease resistance, climate-resilience, and other desirable traits
Results from the 2022 Biodeveloper Survey, summarizing the coding habits and tech-stacks of biologists
FDA approval of first non-hormonal drug for menopause, an NK-3 receptor antagonist that helps normalize body temperature regulation in the brain
A foundation model for single cell RNA-seq called scGPT, providing early evidence for a general-purpose model for a range of single cell tasks including cell annotation, batch normalization, and gene perturbation prediction
Spatiotemporal cell and gene trajectories of human retinal organoid development
What we read
Blogs
New precision-breeding law unlocks gene editing in England [Caccamo, Nature Biotechnology Correspondence, May 2023]
The weight of scientific opinion is clear that the risks from gene-edited crops are no greater than those from conventionally bred ones, as summarized by leading regulatory authorities such as the European Food Safety Authority and Health Canada
Earlier this year, the Genetic Technology Bill became an Act of Parliament in the UK, the first one in decades to enable genetic technologies for agriculture and food production. This precision breeding bill will allow new crop traits to be commercialized, that will bring more nutritious, climate-resistant and disease-resistant crops. This places England alongside Argentina, Australia, Japan, Canada and the US in the list of countries that allow gene editing for crops.
Specifically, the law states that “that new crop varieties or animal breeds that are created using a technology that introduces genetic changes that could have occurred in nature or via traditional breeding methods should not be regulated as genetically modified organisms”. This means that crops where the modification doesn’t introduce foreign DNA will be categorised as ‘precision-bred organisms’ instead of being labelled a GMO.
Recent studies have shown the potential of precision-breeding:
“Wheat varieties that could be suitable for consumers who are gluten intolerant have been created by removing the epitopes from the gliadin proteins in wheat gluten
The ‘Sicilian Rouge’ tomato variety (which has been commercialized in Japan) has been edited to contain high concentrations of γ-aminobutyric acid, a compound that is associated with lowering blood pressure
Scientists have recently used gene editing to increase provitamin D3 in both the fruit and leaves of tomato plants. This, in turn, is converted into vitamin D3 through exposure to UVB light
Roots with steeper angles produce plants that can access water in deeper soil layers, which contributes to the ability of the crop to maintain yields during periods of drought. These traits will be particularly important for growers in conditions such as the summer of 2022 in Europe, when almost no rain was recorded in June and July and temperatures above 40°C were reached for the first time in several northern countries.”
2022 Bio Developer Survey [Cradle Bio x Bits in Bio, May 2023]
The 2022 Bio Developer Survey results are in! The first of its kind, this survey hosted by Cradle Bio asked 160 biotech enthusiasts about their coding habits, career aspirations, and favorite lab equipment. Of the 160 enthusiasts, 20% were wet lab scientists, 70% in silico (dry lab), and 14% claimed to straddle wet and dry lab work environments.
Some more highlights:
87% of wet lab scientists are writing code, and nearly three-fourths are self-taught programmers.
The top programming languages used by biodeveloper respondents are Python, R, Shell, and SQL, and there aren’t any signs of this changing over the next 12 months.
73% of respondents are using machine learning (ML) in their work today, and 82% are interested in applications of ML in bio
The biodeveloper stack is heavy on Amazon Web Services (79%), followed by Google Cloud (45%) and Microsoft Azure (15%). The most popular file formats used by the respondents were CSV/TSV, JSON, FASTA, FASTQ, and BAM/SAM.
In terms of data vizualization, turns out Excel isn’t just for investment bankers, as it remains a favorite of over a third of biodevelopers, following Plotly (45%) and MatPlotLib (63%)
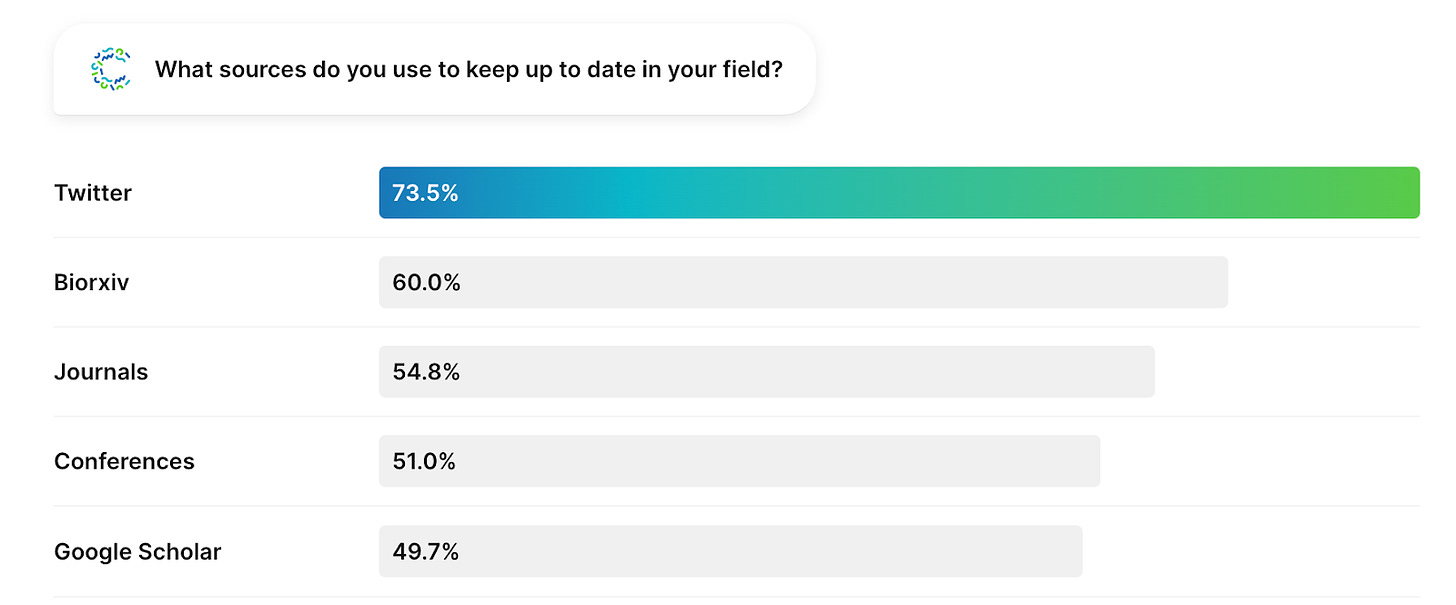
Twitter and pre-print servers continue to be leading sources of information for biodeveloper communities, and ~10% of biodevelopers selected “other” which we will assume means they were Decoding Bio readers 🙂
We are ecstatic about this first-ever biodeveloper survey, and look forward to future analyses that provide additional breakdowns of the computational tools unlocking the future of the bioeconomy.
FDA approves the first nonhormonal menopause drug [Food and Drug Administration, May 2023]
The FDA approved the first nonhormonal drug for menopause last Friday, specifically targeting hot flashes (also known as vasomotor symptoms). Veozah, developed by Astellas Pharmaceuticals, is the first drug on the market that stabilizes neurons that are disturbed when estrogen levels fall, working by binding and blocking the activity of NK3 receptors that are involved in the brain’s regulation of body temperature. The FDA gave Astellas Priority Review for this drug and evaluated two randomized phase three trials that each lasted for a year. Both trials were double-blind and randomized; women on placebo were re-randomized to Veozah after the first 12 weeks, garnering some support for safety. After the course of a year, the drug was deemed generally safe though there are some concerns for liver damage, hence the warnings against using Veozah if the patient has known liver problems. The drug is estimated to cost $550 for a 30-day supply, quite steep given women may have symptoms of menopause for over four years.
NIH Systems for non-human primate research falls short, threatening biomedical research [Ed Silverman, STAT News, May 2023]
Non-human primate data is invaluable in biomedical research given their genetic, behavioral, and physiological similarities to humans. But, we’re in the midst of an NHP shortage given the lack of US infrastructure and resources dedicated towards supporting NHP breeding. Why is this suddenly becoming a problem? Up until the pandemic, we’ve heavily outsourced these studies to other countries, especially China. With the pandemic increasing the number of NHP studies and China’s export ban on the animals, we’ve come to realize that very little infrastructure exists for the US to support these studies—and there are long wait times given the demand. And while NHP data is valuable in biomedical research, bolstering NHP resources is a contentious problem with two opposing sentiments—animal rights activists who want to move away from domestic production of monkeys for research entirely—and the biomedical research groups that claim NHP studies are currently irreplaceable as we require studying a living organism to advance human treatments. While several alternatives to animal testing are emerging (organ on a chip, human model systems, etc), we haven’t yet found a model that is as validated as non-human primates.
President’s Orders: Executing a Bioeconomy [Vega Shah and Shelby Newsad, May 2023]
The Biden-Harris administration recently issued the Executive Order on Advancing Biotechnology and Biomanufacturing (EOABB), with an aim to stimulate activity in the biotechnology and biomanufacturing sectors by committing to an investment of $2B. Vega and Shelby unpack what the executive order means for the bioeconomy. A strategic framework published in March 2023 outlines key goals on the initiative, including accelerating the development of new biotech products, building a robust domestic biomanufacturing industry, fostering innovation and entrepreneurship, developing a skilled workforce, and promoting responsible and ethical biotech practices.
While much of the executive order is fluffy rhetoric at this point, a couple of tangible proposals are worth highlighting. First, the aggregation of multi-omic data across industries including healthcare, climate, and food/ag. While commendable, how this will be enforced and executed is unclear. The EO also emphasizes biobased product procurement, which will require government staff to be trained in biobased product purchasing. To address this, the plan includes an educational plan for upskilling the workforce to meet these demands.
Academic papers
scGPT: Towards Building a Foundation Model for Single-Cell Multi-omics Using Generative AI [Cui et al., bioRxiv, May 2023]
Foundation models, a term coined in a 2021 paper by a group of Stanford researchers, are large, usually transformer-based models trained on massive unlabelled datasets using self-supervised learning. When trained at sufficient scale, these models demonstrate emergent behaviors, capable of generalizing to many different tasks. For example, LLMs like ChatGPT trained to perform a simple task like next word prediction results in many emergent capabilities such as the ability to do math, follow instructions, answer questions, and summarize passages.
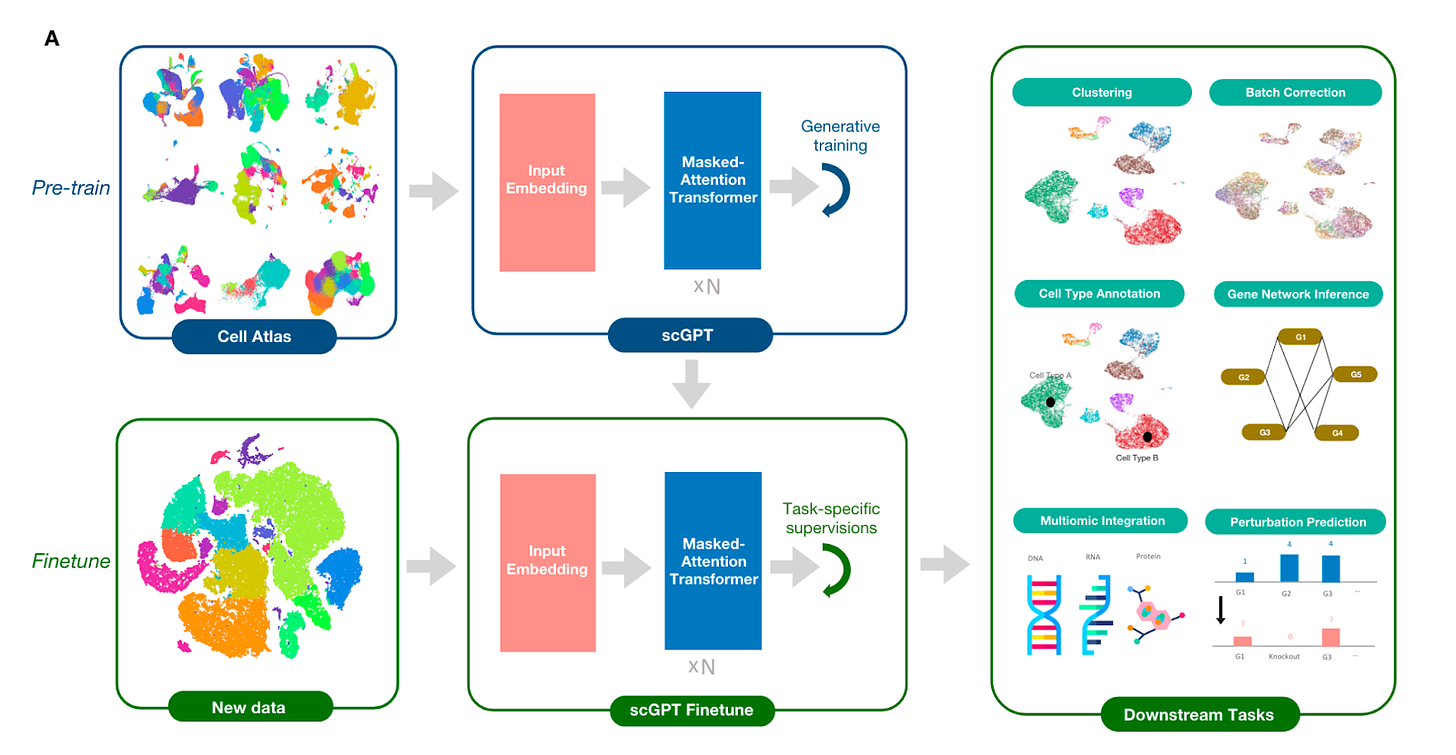
Bo Wang’s lab at the University of Toronto recently published a new foundation model for bio called single-cell GPT (scGPT). In this paper, a generative pre-trained (GPT) model was trained using masked token prediction (i.e., randomly masked tokens in the input data are predicted), similar to LLMs like ChatGPT. A pre-trained foundation model was generated by training a transformer-based architecture on a self-supervised task involving predicting gene expression based on partial gene expression data ("gene prompts"), or whole genome expressions given an input cell condition ("cell prompts"), analogous to how ChatGPT learns to predict the next word in a sentence. The model successfully generalized to new tasks via fine-tuning (i.e., adapted to new tasks using a much smaller training set), such as:
Batch correction (correcting variation in scRNA-seq results due to technical factors such as differences in workflow, rather than underlying biology)
Pancreas cell type annotation, outperforming other top annotation models
Multi-omic integration of gene expression, chromatin accessibility, and protein abundance data to via joint representation learning to improve cell clustering and batch correction performance
Gene perturbation prediction - scGPT could predict whole genome gene expression changes resulting from single gene perturbations in Perturb-seq datasets (high-throughput characterization of transcriptional changes in response to CRISPR gene inactivation)
scGPT was trained on 10 million blood and bone marrow cells, yet there are publicly available single cell datasets for many other cell types. It will be interesting to reproduce and extend the model capabilities using a larger and more biologically diverse dataset. One big question in the field of foundation models for bio is what is the best pre-training task for engineering general-purpose models. Masked token prediction like next word prediction works well for sequence data like natural language, but most biological data lacks linear, sequence-based structure. This paper had a clever workaround, but an important line of research going forward will be the systematic exploration of different pre-text/pre-training tasks.
Multimodal spatiotemporal phenotyping of human retinal organoid development [Whale et al., Nature Biotechnology, 2023]
Single-cell sequencing and image-based measurements have allowed developmental biologists to reconstruct cell state trajectories, offering new insights into the differentiation dynamics across lineages and spatial domains.
When combined with human stem cell-derived organoids, these technologies can help elucidate the molecular definition of cell states, how they relate to tissue structure and their morphological development. The combination of organoids and multimodal data at spatial and temporal dimensions can allow for the prediction of cell state changes under different perturbations. However, integrating multimodal data across spatial scales is a major challenge, especially within human embryonic sample-derived organoids which have substantial heterogeneity.
The authors of the paper established an experimental pipeline, using 4i (iterative indirect immunofluorescence imaging) on histological sections of retinal organoids, alongside a computational approach for inferring spatial developmental dynamics for proteins maps for heterogeneous organoids. They combined it with scRNA-seq and scATAC-seq covering 6-46 weeks of development, which can reconstruct differentiation trajectories into each of the major neuronal and glial lineages.
By integrating the above datasets, they developed the “first-of-its-kind digital representation of human retinal organoid development”. They add: “the digital organoid map can be used to explore spatial interactions over time and predict gene regulatory modules underlying retinal neurogenesis”
Efficient evolution of human antibodies from general protein language models [Hie et al., Nature Biotech, April 2023]
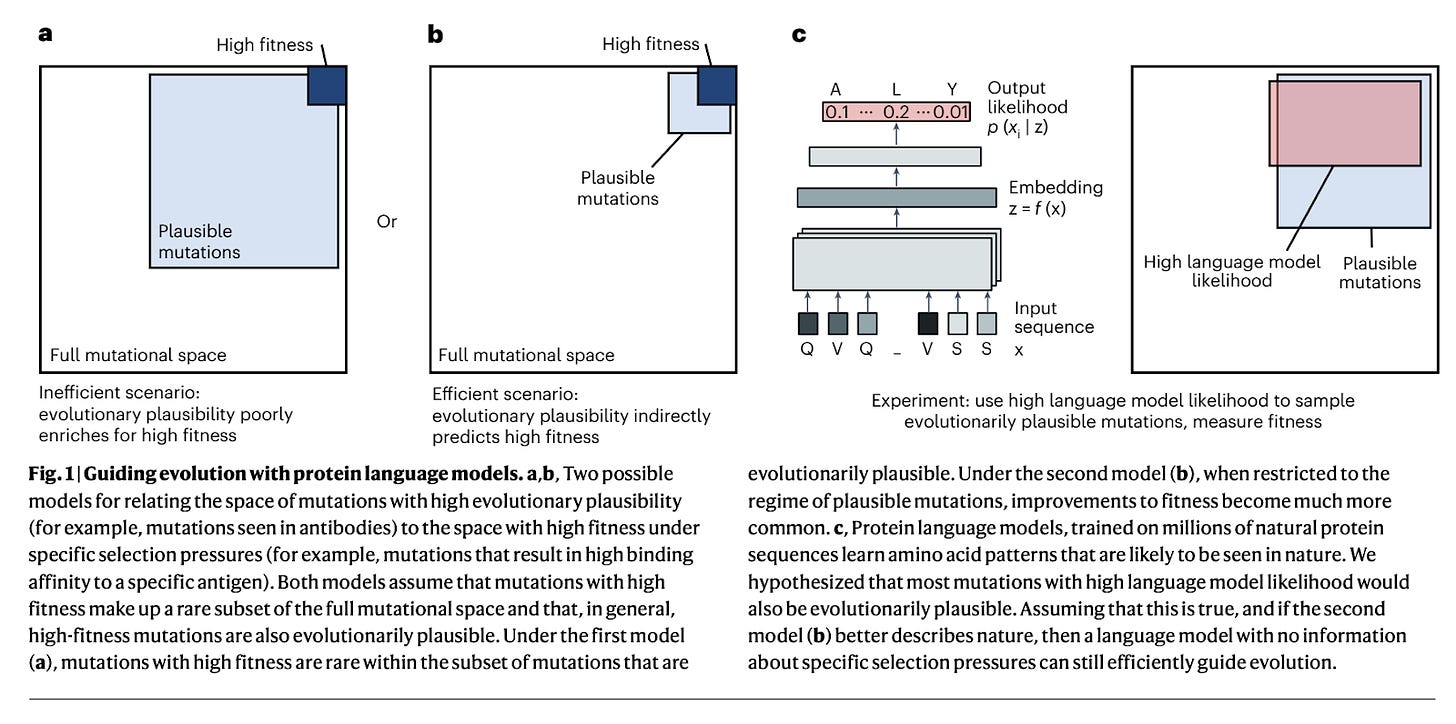
This paper discusses the use of natural evolution as a guide for AI-enabled evolution in the context of protein engineering. Hie et al. found that general protein language models can effectively evolve human antibodies by suggesting plausible mutations, even without prior knowledge of the target antigen, binding specificity, or protein structure. Through language-model-guided affinity maturation, the team demonstrated improved binding affinities of seven clinically mature antibodies including those representing antigens for SARS-CoV-1 and 2. Additionally, the language models were capable of guiding efficient evolution across diverse protein families and selection pressures, including antibiotic resistance. Perhaps most impressive about this work is the team’s ability to improve already clinically mature antibodies while demonstrating best-in-class computational efficiency (1 second per antibody prediction!) relative to historical methods.
Unlocking de novo antibody design with generative artificial intelligence [Shanehsazzadeh et al., bioRxiv, May 2023]
Continuing down the road of computational antibody design, Absci’s machine learning team just put out this updated preprint detailing their zero-shot learning (i.e., adapting a pre-trained model to a new downstream task without re-training the model with additional task-specific training data) methods to generate completely de novo antibodies. This means they are using generative models to design entirely new antibodies without fine-tuning and training those models with follow-up optimization. More specifically, the antibody binders generated are from models that have never seen binders to the antigen in question before. They tested their models against three separate targets and used surface plasmon resonance (SPR) to characterize binding of 421 binders, finding three variants bound more tightly to the target (HER2) than existing therapeutic monoclonal antibody trastuzumab, which you may know as the breast cancer drug Herceptin. Encouragingly, the sequences of these antibodies were quite diverse, showing the models are generating binders that span a wide-space and don’t optimize for any particular motif. Downstream antibody developability is a challenging problem for drug development, and zero-shot learning is well on its way to addressing some of these challenges.
If you enjoyed the last two papers and are still curious about computational antibody design, check out this recent piece by Derek Lowe, “Computing Our Way to Antibodies.”
What we listened to
Notable Deals
AAV producer NewBiologix emerges from stealth with $50M Series A
Gilead acquires San Diego startup for early-stage cancer, immune drugs
NewLimit, co-founded by Coinbase CEO Brian Armstrong, raises $40M to extend life
New Eli Lilly-backed neurological disease biotech Nido unveils with $109M
FTC sues to block Amgen’s $27.8B deal for Horizon
Flagship’s newest startup is imitating nature to make new drugs
In case you missed it
Google Cloud launches A.I.-powered tools to accelerate drug discovery, precision medicine
What we liked on Twitter
Events
Field Trip
Did we miss anything? Would you like to contribute to Decoding Bio by writing a guest post? Drop us a note here or chat with us on Twitter: @ameekapadia @ketanyerneni @morgancheatham @pablolubroth @patricksmalone