BioByte 031: the AI for chemistry revolution, safety in gene therapy, AlphaFold's structural shortcomings, mapping antisense oligos in the brain
Welcome to Decoding Bio, a writing collective focused on the latest scientific advancements, news, and people building at the intersection of tech x bio. If you’d like to connect or collaborate, please shoot us a note here or chat with us on Twitter: @ameekapadia @ketanyerneni @morgancheatham @pablolubroth @patricksmalone. Happy decoding!
A few of us are floating around SynBioBeta this week… if you’re around, let’s link up!
Another big week for bio and we’ve got you covered. Short on time? Here are some of the highlights:
We’re on the cusp of an AI revolution in chemistry. In silico de novo synthesis and optimization are unlocking entirely new classes of compounds—not unlike what AlphaFold did for protein synthesis… but we desperately need more training data that includes both positive (i.e., produced a desired molecule) and negative (i.e., did not produce a desired molecule) outcomes…
Researchers uncover new information about the safety of gene therapies after studying the story of Terry Horgan, a 27-year-old man with Duchenne muscular dystrophy who was the first person to receive a designer CRISPR therapy.
GeneGPT offers a new method for teaching LLMs to use the APIs of the National Center for Biotechnology Information (NCBI) to answer genomics questions.
New research leveraging single-cell sequencing reveals nuances of how anti-sense oligonucleotides (ASOs), or short fragments of nuclei acid polymers with high therapeutic relevance, affect certain cell types in the brain that may be differentially implicated across diseases.
What we read
Blogs
For chemists, the AI revolution has yet to happen [Nature Editorial, May 2023]
We talk a lot about GPT in medicine and biology, both of which have come a long way in just a few months. AI in chemistry, however, has been lacking—primarily bottlenecked by one looming problem: lack of accessible data. AI in chemistry can look like de novo synthesis and optimization of entirely new classes of compounds—not unlike what AlphaFold did for protein synthesis, which you can argue was an advancement in AI for chemistry itself. The advantage AlphaFold had that other areas of chemistry do not, however, is an accessible large database of thousands of structures that can be used to train a model. Open source data, automated chemical experiments to gather more data points, and generating simulated data are all potential solutions to this problem. But the authors bring up a good point:
“The best possible training sets would also include data on negative outcomes, such as reaction conditions that don’t produce desired substances. And data need to be recorded in agreed and consistent formats, which they are not at present.”
In a related piece by Andrew White in Nature Reviews Chemistry, he proposes that the future of chemistry is language! In this brief editorial, Dr. Andrew White outlines why large language models (LLMs) are going to transform how we approach complex tasks in chemistry. LLMs are already impacting modern reaction synthesis planner tools and molecular property explanation, and Dr. White argues that LLMs will find their way into every aspect of the field. Some of his predictions about other applications include:
Using LLMs to make IUPAC names, or even common names, as inputs to molecular prediction tasks
Writing computational chemistry code, lowering the barrier of entry for writing density functional theory (DFT) input files or analyzing protein structure
Semantic literature search for chemistry
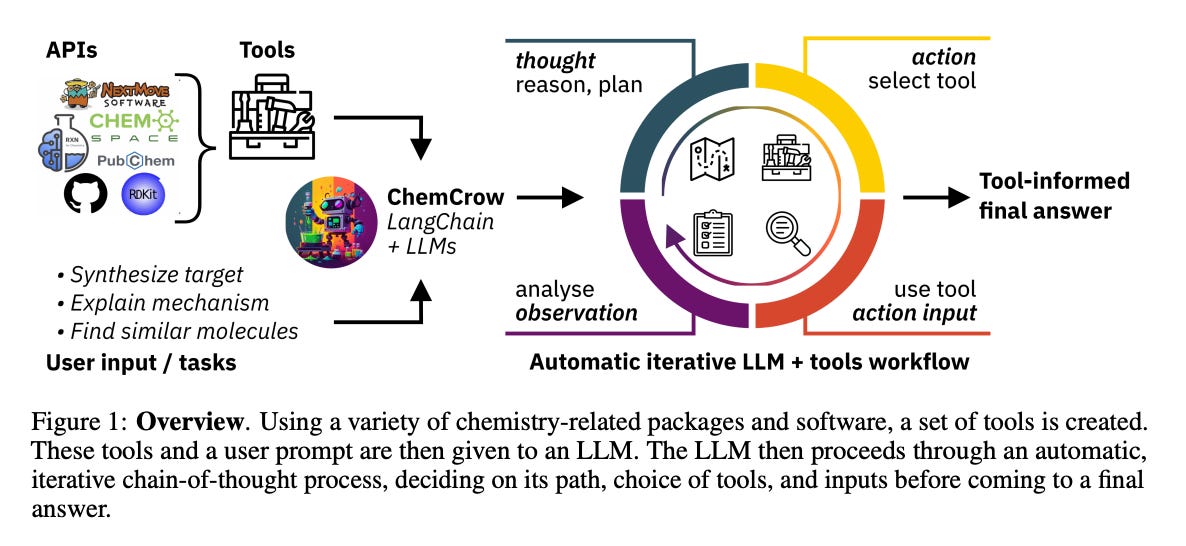
The piece relates deeply to Dr. White’s academic work, which you can follow here and on Twitter. Recently, Dr. White and his team introduced ChemCrow, an LLM chemistry agent designed to accomplish tasks across organic synthesis, drug discovery, and materials design.
Patient who received custom gene therapy likely died from immune reaction, not CRISPR, paper says [Endpoints, 2023]
For those of you following the gene therapy field closely, Terry Horgan’s name is familiar to you. He was a brave 27-year-old man with Duchenne’s muscular dystrophy (DMD) who passed away in October of 2022 after receiving a bespoke CRISPR activation therapy for his DMD. The cause of Terry’s death – eight days post-treatment – has been a subject of investigation. Shortly after administration, Terry developed signs of mild cardiac dysfunction, and a pericardial effusion before acutely decompensating, developing signs of symptoms of acute respiratory distress syndrome (ARDS) and progressing to cardiopulmonary arrest. Although he was placed on extracorporeal membrane oxygenation (ECMO), Terry passed away 8 days post-treatment secondary to multiorgan failure and severe neurological injury.
What was the underlying reason for the rapid deterioration – was it the actual editing? Was it the CRISPR components? An autopsy was carried out and revealed that Terry had minimal amount of CRISPR production in the liver, and none in the extrahepatic organs. However, analyses demonstrated he had a significant innate immune response to the AAV, resulting in capillary leak syndrome (where plasma, proteins, etc. leak out of blood vessels) and resulted in his pericardial effusion and ARDS.
Taken together, this story sheds light on the continued discussion about the safety of gene therapies. Reducing immunogenicity of vectors (and components), or even more specifically abrogating the immune response peri-treatment may be crucial for ensuring safety across patients. For all of you reading this… there is always a patient on the other side. Let’s never forget that. Thank you to all patients who bravely put their health on the line for others.
Supreme Court rules invalid Amgen’s patent on cholesterol-lowering drug [Eric Fraser, Scotus Blog, 2023]
In a recent case, the Supreme Court ruled against Amgen, a pharmaceutical company that had sued Sanofi for violating its patent on antibodies that reduce LDL cholesterol. The court deemed Amgen's patent invalid because it lacked sufficient information for others to create and utilize the entire range of antibodies covered by the patent. Justice Neil Gorsuch, writing for a unanimous court, explained that Amgen's patent covered a broad class of antibodies, potentially encompassing millions of antibodies, and failed to provide proper enablement. While a patent doesn't have to detail every embodiment within a claimed class, it must offer examples and specific characteristics to enable others to make and use what the patent claims. This ruling sets a precedent for the difficulty in satisfying the enablement requirement for broad-genus patents in fields like pharmaceuticals, chemicals, and biotechnology.
“The more one claims, the more one must enable.”
In a related opinion piece published this week by Dennis Crouch, Law Professor at the University of Missouri School of Law, he explains:
The outcome of the Amgen v. Sanofi case, while significant, may not come as a surprise to those familiar with the evolution of patent law. In recent years, there has been a growing emphasis on promoting innovation and competition by limiting the scope of patent monopolies. This approach recognizes that patents should strike a balance between incentivizing inventors to disclose their inventions and ensuring that the public can freely build upon existing knowledge. Rather than granting broad and far-reaching patent rights, the trend has been towards encouraging more focused and narrowly tailored claims. This shift reflects a recognition that patents are more palatable when they offer bite-sized protection rather than providing a singular and overpowering monopoly. The ruling in the Amgen v. Sanofi case aligns with this approach, reinforcing the notion that patents should enable the creation and use of the claimed inventions while still allowing for competition and further advancements in the field.
The under-appreciation of CHIP [Eric Topol, May 2023]
CHIP, or clonal hematopoiesis of indeterminate potential, refers to the expansion of a genetically distinct sub-population of myeloid blood cells or other progenitor cells. CHIP is a biomarker for a range of diseases including blood cancer, heart disease, blood clots, and other chronic diseases. Over 10% of people over the age of 70 have CHIP, and the proportion increases with advancing age. There are several characteristic driver mutations of CHIP, such as TET2 and DNMT3A which are involved in DNA methylation and other epigenetic modifications. Given the research that has emerged over the last 10+ years, Eric argues for the prioritization of CHIP as a biomarker with translational potential. We are beginning to see new biotechs being started in this space. TenSixteen Bio, a Foresite and GV startup, a precision medicine company developing therapeutics targeting CHIP. TenSixteen is building a multi-omics and clinical data platform on CHIP, and is developing novel diagnostics CHIP assays. The goal is to discover novel CHIP therapeutic targets, risk-stratify patients according to CHIP profile and mutational burden, and develop precision-approaches for treatment using novel biomarkers.
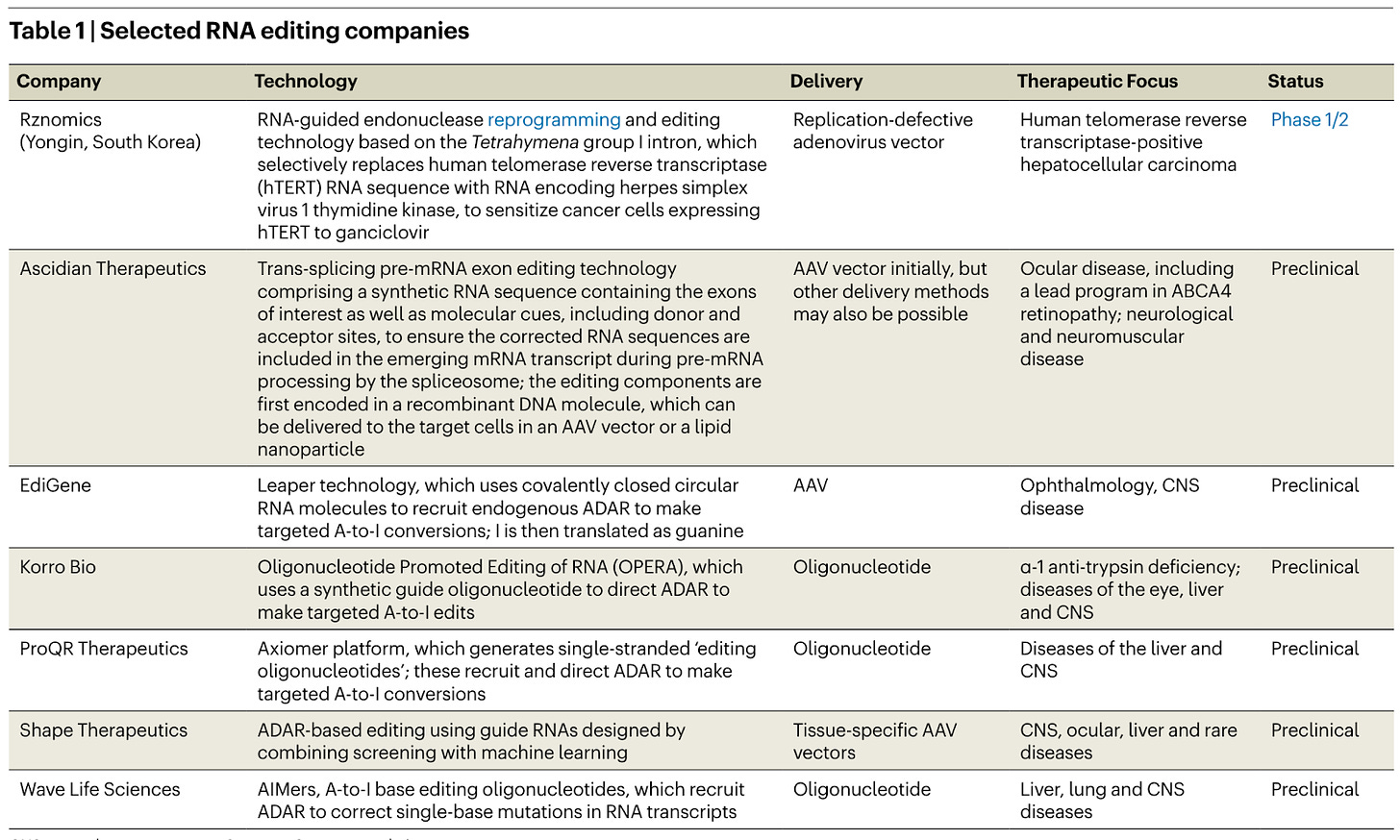
Shoot the messenger: RNA editing is here [Sherdian, Nature Biotech News, 2023]
By editing RNA instead of DNA, one can create transient changes, leading to safer therapeutics. Companies developing RNA editing therapeutics that harness the endogenous enzyme system of ADARs have seen major partnerships with Pharma (ProQR-Eli Lilly, Wave-GSK and Shape-Roche). Wave’s WVE-006 program aims to be the first to the clinic, treating antitrypsin deficiency (AATD). ADARs are analogous to the DNA base editors, switching one nucleotide for another.
Ascidian uses a different method. It exploits the spliceosome, which supports pre-mRNA editing within the nucleus, to trans-splice correct versions of mutated genes.
“ADAR is like fixing typos. The splicing approaches are rewriting paragraphs,” says Sullenger
This approach is suited to “correcting mutations in large genes that exceed the packaging limit of AAV vectors and to replacing large stretches of genetic code in which multiple disease-causing mutations are present in the patient population.”
Academic papers
GeneGPT: Augmenting Large Language Models with Domain Tools for Improved Access to Biomedical Information [Jin et al., arXiv, 2023]
GeneGPT is a new method for teaching LLMs to use the APIs of the NCBI to answer genomics questions. GeneGPT surpassed the performance of general and domain-specific LLMs such as Bing, GPT-3, BioGPT and BioMedLM. The key novelty is that GeneGPT uses NCBI Web APIs calls for in-context learning. By prompting GeneGPT with API documentation and API demonstrations, the LLM then carries out API calls itself (through OpenAI’s codex), retrieving information more accurately than with an LLM trained on the data set itself. Pretty neat.
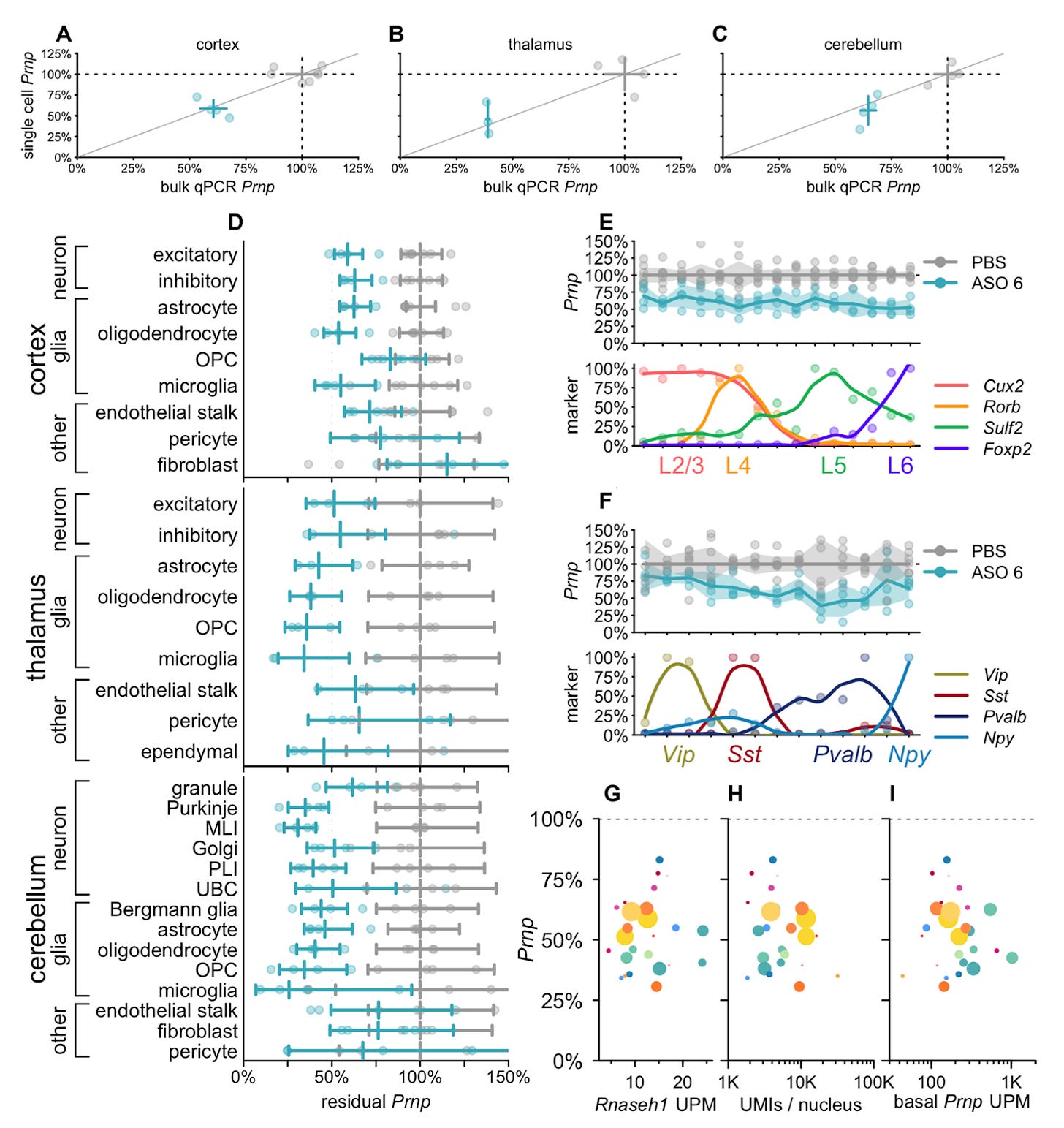
A single-cell map of antisense oligonucleotide activity in the brain [Mortberg et al.,. Nucleic Acids Res., 2023]
Why it matters: Antisense oligonucleotides (ASOs) are short fragments of nucleic acid polymers with the potential to treat several diseases by modifying transcription dynamics. In recent years, they have demonstrated marked potential in the ability to treat neurodegenerative disease. However, to date, their fate in the CNS has remained poorly understood, and is critical to understanding their utility as ASOs are increasingly used across neurological disease. In this work, Mortberg et al., use single cell RNA sequencing to understand ASO activity in the brain – from localization to knockdown dynamics.The authors treated mice (intracerebroventricular) and NHPs (intrathecal) with RNase H1 ASOs previously characterized to extend survival in prion infection: Prnp and Malat1. Subsequently, they used single-nucleus transcriptomics to identify ASO dynamics across tissue. Their results were striking: they found that ASOs lower target RNA across all cell types in tissue that take up the drug in both mice and NHPs; additionally, the amount of target RNA reduction is different across cell types (e.g. Purkinje Cells demonstrated 17% residual, while microglia had 57%), and duration of action is also different (e.g. microglia nearly recovered expression by 12 weeks while excitatory neurons were steady). Of note, the authors were unable to explain differences in knockdown between cell types by target expression, UMI count (proxy for # of transcripts/ cell size), and RNAse H1 levels. This remains an open area of investigation.
This work is incredibly important as it provides a foundation for scientists to understand the nuances of how ASOs affect certain cell types that may be differentially implicated across diseases. Furthermore, it lends credence to the hypothesis that a decrease in certain biomarkers (such as PrP levels in the CSF) may truly reflect a true lowering of said target in the cells that actually matter.
Sites of transcription initiation drive mRNA isoform selection [Carlos Alfonso-Gonzalez et al., Cell, 2023]
Why it matters: Every gene in our DNA has a specific starting and ending point. Defining these endpoints accurately in relation to transcription is essential for ensuring the production of functional proteins. While extensive research has focused on where genes begin, determining the termination site of a gene has been more challenging. This work sheds light on the intricate interplay between transcrription start sites and end sites, demonstrating a causal relationship and an impact on mRNA diversity. The findings have implications for a wide range of biological processes and can enhance our understanding of genetic regulation and the development of different tissues and organisms.Researchers from the Max Planck Institute of Immunobiology and Epigenetics have made a significant discovery regarding the relationship between transcription start sites (TSSs) and transcription end sites (TESs) in genes. They found that for most genes, the site of transcription start determines the site of transcription end, which has important implications for cell identity and functionality. This phenomenon is conserved across species and provides insights into how mRNA molecules are generated, adding a new layer of complexity to the study of the genome.
Methodologically, the team leveraged a modified version of next-generation sequencing to read and analyze individual messenger RNA (mRNA) molecules from various model systems, including Drosophila tissues and human cerebral organoids. By optimizing long-read sequencing methods, they were able to obtain full-length mRNA information, which provided unprecedented insights into the transcription process of genes. This analysis allowed the team to disentangle the relationship between TSSs and TESs and uncover the role of dominant promoters in shaping the RNA landscape and tissue identity. Additionally, they performed sequence conservation analysis to explore the evolutionary aspects of TSSs and TESs, providing further support for the importance of these gene extremities in maintaining animal fitness.
AlphaFold predictions are valuable hypotheses, and accelerate but do not replace experimental structure determination [Terwilliger et al., BioRxiv, May 2023]
Why it matters: A recent study comparing AlphaFold-generated protein structures to experimentally-derived crystal structures found important inconsistencies, highlighting the importance of predicting in silico and validating in vitro/vivo. To what extent can protein structure predictions from systems like AlphaFold substitute for experimental structure determinations? This paper compares high-confidence protein structure predictions from AlphaFold to experimentally-derived crystal structures. An important limitation of protein folding algorithms like AlphaFold is that they do not take into account the presence of ligands, covalent modifications, or environmental factors, all of which can dynamically influence a protein’s structure. While in general AlphaFold predictions were found to be accurate overall, many parts of AlphaFold predictions were found to be inconsistent with experimental data. The authors propose that AlphaFold predictions should be treated as hypotheses that should be validated via experiment. In other words: predict in silico, validate in vitro/vivo.
What we listened to
Notable Deals
Siren Biotechnology Launches to Pioneer Universal AAV Immuno-Gene Therapy™ for Cancer
Belgian biotech Dualyx enters Treg field with $44M for 2024 clinical entry
Myeloid Therapeutics raises $73M, bringing in ARCH ahead of in vivo program entering the clinic
ReNAgade Therapeutics launches with $300 million, joint venture to expedite RNA drug development
Known Medicine acquired by Pathos AI
In case you missed it
Advancing structural biology through breakthroughs in AI [Aithani et al., Current Opinion in Structural Biology, 2023]
What we liked on Twitter
Events
Field Trip
It’s almost hip-hop barbeque season… we’re throwing it back! Enjoy some Ja 🎶
Did we miss anything? Would you like to contribute to Decoding Bio by writing a guest post? Drop us a note here or chat with us on Twitter: @ameekapadia @ketanyerneni @morgancheatham @pablolubroth @patricksmalone