BioByte 033: fragmentomics, mainstream gene therapies, the immune system and psychosis, herpes zoster vaccination and dementia, ProteinChat, human disease models
Welcome to Decoding Bio, a writing collective focused on the latest scientific advancements, news, and people building at the intersection of tech x bio. If you’d like to connect or collaborate, please shoot us a note here or chat with us on Twitter: @ameekapadia @ketanyerneni @morgancheatham @pablolubroth @patricksmalone. Happy decoding!
In a hurry and in between meetings at BIO? Here’s what we covered this week:
Getting cell and gene therapies to patients: what will it take for gene and cell therapies to become mainstream?
Fragmentomics: by examining the pattern of DNA fragment amounts and sizes in the blood, researchers have developed a promising blood test to detect liver cancer, even in its early stages
The immune system and psychosis: how inflammatory disorders can lead to disorders indistinguishable from schizophrenia
Herpes zoster vaccination prevents dementia in 20% of people
Predictions in network biology: a foundation model pre-trained on 30M human single-cell transcriptomes learns representations for hierarchical gene networks, enabling data-efficient generalization
ProteinChat: a prototype GPT model specialized in learning and understanding protein structures.
Human disease models in drug development: new bioengineered human disease models can accelerate drug development by exhibiting a higher level of clinical mimicry
What we read
Blogs
Addressing the Challenges of Getting Gene and Cell Therapies to Patients [Helen Albert, Inside Precision Medicine, May 2023]
What will it take for gene and cell therapies to become mainstream? In this piece, Helen Albert describes key challenges facing gene and cell therapies in the face of an active discovery and development market: 28 cell and gene therapies approved by the FDA to date, and 6 new therapies approved in the U.S. or Europe in 2022 alone.
The article covers challenges related to:
Patient identification and diagnostics
Rare genetic diseases have been a key focus area for gene therapy companies. According to the Undiagnosed Disease Network, 25 million Americans suffer from rare disorders. Yet according to the NIH, only 500 of 7,000 rare diseases have approved treatments. Put simply, improving the diagnosis of rare diseases will help improve access to gene therapies.
Newborn screening is one diagnostic modality that is increasing in popularity, but we face a Catch-22: “if there is not already an approved therapy for a condition, it will not usually be included in standard heel prick blood testing of newborns.” For example, in the case of Angelman Syndrome, there are 500,000 kids predicted to have Angelman syndrome globally, yet we are aware of fewer than 10,000 today.
Complexity of therapy
Current cell therapies, such as autologous CAR T cell therapies (a personalized immunotherapy approach that involves genetically modifying a patient's own T cells to target and kill cancer cells), are challenging to manufacture and thus often take a long time to reach patients in need. To address these challenges, some companies are developing “off the shelf” allogeneic (donor) cell therapies, with the hopes of reducing manufacturing complexity and time; however, risks of rejection present barriers for this approach compared to autologous cell therapies. One company, called Orgenesis, is developing technology and processes to allow cell therapies to be prepared on-site/near-site.
Pricing and payment
Gene and cell therapies are notoriously expensive, with many price tags in the millions of dollars, which is enough to bankrupt a self-funded employer. As a result, these therapies are increasingly carved out of value-based care and other cost-containment models in healthcare delivery. There continues to be a robust discussion of new models, including:
Encouraging health plans to cover cell and gene therapies include paying in installments rather than all at once
Paying for performance (i.e., reimbursement is returned if the therapy doesn’t work)
Re-insurance, where a payer itself is insured against having to pay out for a cell or gene therapy
Risk-pooling model where payers join together to spread the risk
Can the New “Omics” on the Block Find Liver Cancer in Blood? [Nadia Jaber, National Cancer Institute, May 2023]
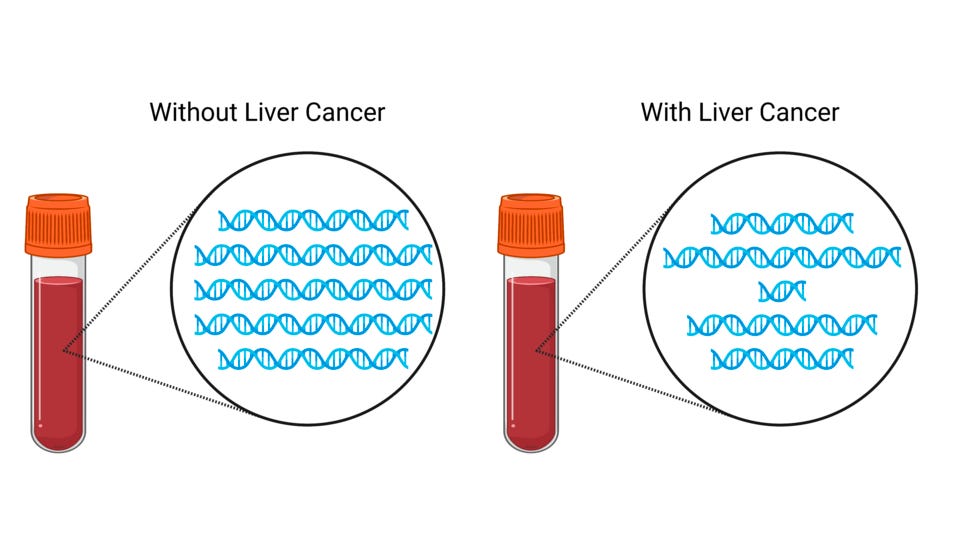
Liver cancer is the third leading cause of cancer-related deaths globally, but early detection significantly improves survival rates. The current tests for liver cancer are inadequate, inaccessible, and expensive. Researchers have developed a promising new blood test using fragmentomics technology to detect liver cancer, even in its early stages.
Scientists have discovered that DNA fragments shed by both healthy and cancerous cells in the bloodstream can be used as markers to detect cancer. While most liquid biopsy tests focus on genomics or epigenomics, which analyze DNA mutations or chemical tags on DNA, fragmentomics examines the pattern of DNA fragment amounts and sizes in the blood.
In the case of liver cancer, researchers discovered that the size of DNA fragments in the blood of individuals with liver cancer varies significantly, unlike the consistent size of fragments in the blood of those without the disease. A recent application of fragmentomics technology called DELFI, developed by Dr. Victor Velculescu at Johns Hopkins, demonstrated impressive accuracy in identifying liver cancer patients with an AUC of 0.98.
So where do we go from here? Experts believe that fragmentomics approaches boast advantages over liquid biopsies for cancer detection:
Fragmentomics show immense potential to develop pan-cancer diagnostics
Unlike mutation- and methylation-based tests, which rely on rare cancer-related changes in the blood, fragmentomics requires less intensive scanning, reducing costs
Fragmentomics necessitates a smaller blood sample compared to other liquid biopsy methods
A catatonic woman awakened after 20 years. Her story may change psychiatry [Richard Sima, Washington Post, 2023]
This remarkable article tracks two young women, April and Devine, that seemingly from one day to the next started showing signs of schizophrenia or schizoaffective disorder. Sander Markx, Director of Precision Psychiatry at Columbia University, and his team, established the relationship between neuroinflammation and April’s and Devine’s psychoses. After an intense course of immunosuppressive drugs, both of them “awakened” from their conditions and now are living a life outside psychiatric institutions, writing poetry and tasks that were unthinkable to them.
This shows, amongst other research, the promise of the linkage between inflammation and psychology, and the urgency to implement the cheap diagnostic tests to discard potentially readily treatable conditions in patients with psychosis. This is also a potential avenue to develop more targeted therapeutics than sledgehammer steroids/chemotherapeutic agents for immunosuppression or antipsychotic drugs.
A spotlight on shoddy offsets [Climate Tech VC, May 2023]
The voluntary carbon market is a marketplace for businesses and individuals to purchase carbon offsets to reduce their own greenhouse gas emissions. These purchases are not required by regulation, but are instead made voluntarily to meet corporate sustainability goals, comply with green certification programs, or just as a part of a personal commitment to reducing climate impact. Verification companies like Vera are supposed to verify that projects and companies that sell carbon credits actually lead to a reduction of greenhouse gasses. The Guardian recently reported that 93% of the credits that Chevron bought, many of which were validated by Vera, were worthless. The CEO of Vera has stepped down as a result.
Large companies that have genuine net-zero emission commitments like JPMorgan and Microsoft are now purchasing more expensive, higher-quality credits. Going forward, the industry needs better measurement, reporting, and verification (MRV) technologies to validate the quality and integrity of projects to keep carbon credit sellers and validators accountable.
Academic papers
Causal evidence that herpes zoster vaccination prevents a proportion of dementia cases [Eyting et al., medRxiv, 2023]
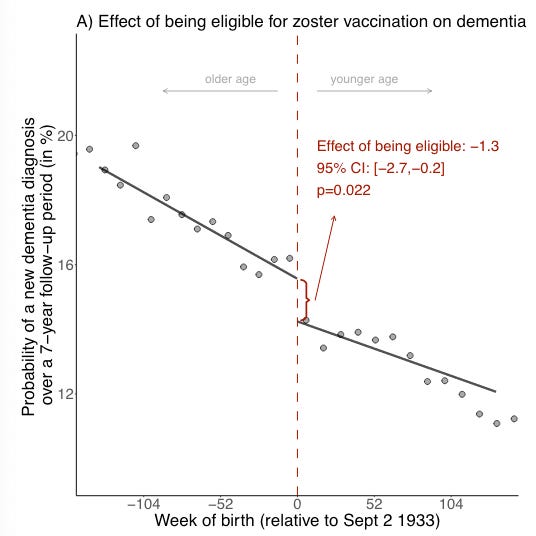
Why it matters: the etiology of dementia is still unclear despite the decades of research and hundreds of clinical studies. This study shows causal evidence between herpes zoster for shingles vaccination and a 20% reduction in the occurrence of dementia. This unique study demonstrates, through natural randomisation, that the herpes zoster vaccine reduces the occurrence of dementia. The authors used the fact that the eligibility for the vaccine in Wales was determined based on the exact birth date of individuals (starting Sept 1st 2013, those born on or after Sept 2nd 1933 were eligible while those born earlier were not).
By using a cohort of individuals that were not eligible for the vaccine as they were born a week earlier, the authors mimicked a randomized clinical trial. They found that over a seven-year follow up period, the probability of a new dementia diagnosis for an individual vaccinated with the herpes zoster vaccine reduced by about 20%
Transfer learning enables predictions in network biology [Theodris et al., Nature, May 2023]
Why it matters: A foundation model pretrained on 30M human single-cell transcriptomes learns representations for hierarchical gene networks, enabling data-efficient generalization via fine-tuning to a range of downstream tasks, including the discovery of a novel candidate therapeutic target for cardiomyopathies. Mapping gene networks, a collection of regulatory elements in a cell that interact to control biological processes, requires large amounts of transcriptomic data. In many applications of biomedical importance (e.g., discovering novel therapeutic targets or biomarkers of treatment response), access to sufficient data is often a bottleneck. To address this challenge, the authors leveraged transfer learning (an important feature of foundation models), or training a model on one task (often with plenty of readily available training data) and then fine-tuning the pre-trained model on a different task with more limited training data. The concept is that the pre-trained model is able to leverage prior learning from the pre-text task to more effectively solve new tasks.
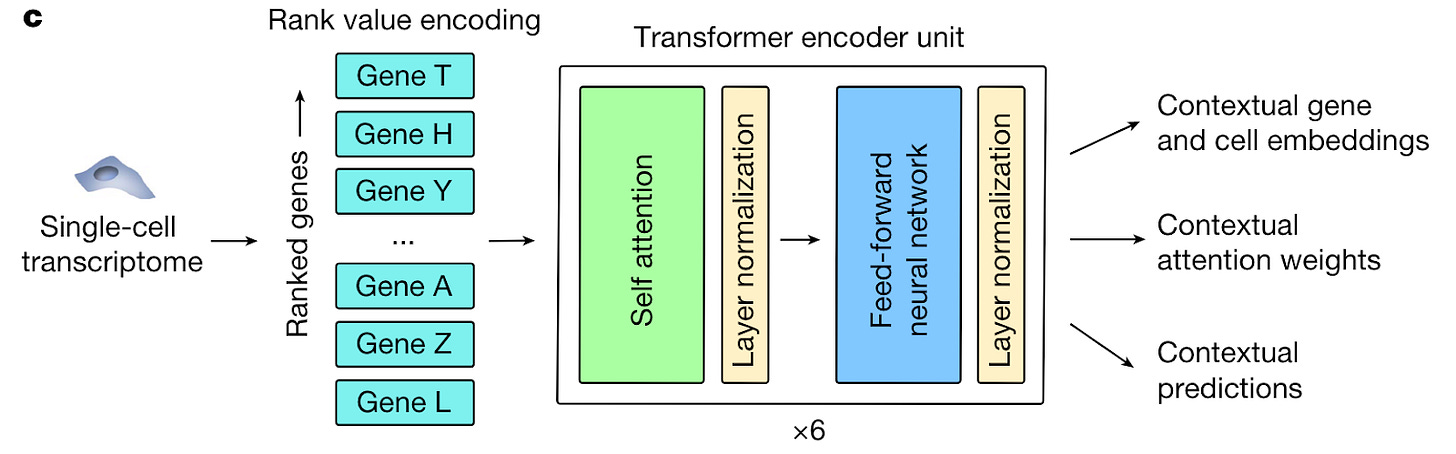
A transformer-based model called Geneformer was pre-trained on 29.9M human single-cell transcriptomes from a range of tissue types. An important innovation was the pre-training task. The transcriptome of each cell was rank ordered according to each gene’s expression, normalized by their expression across all cells in the training set (figure below). This depriortizes universally expressed housekeeping genes that are expressed across most cell types, while strengthening the signal of transcription factors which are expressed at lower levels, but are better predictors of cell state. Like other foundation models, the pre-training task is masked token prediction - genes are randomly masked, and the algorithm learns to predict the identity of the gene in the masked position in a particular cell state.
The model was then fine-tuned and successfully generalized to a range of downstream tasks such as gene dosage sensitivity and chromatin dynamics predictions. Most interestingly, analysis of attention weights of the model (a method for visualizing how a transformer attends to different parts of the input data used to understand how to the model processes data) showed that the model learned gene network hierarchies, preferentially learning representations for central nodes in the network such as central regulatory nodes. In silico pertubration modeling in cardiomyopathic cells discovered a novel therapeutic target by selectiving inhibiting or activating in silico specific genes, and observing whether the overall cell embeddings were shifted back to a healthy cell state. One identified gene, Titin (TTN), was experimentally validated in an iPSC disease model.
ProteinChat: Towards Achieving ChatGPT-Like Functionalities on Protein 3D Structures [Guo et al., TechXiv, May 2023]
Why it matters: Natural language interfaces powered by LLMs are unlocking novel ways of interfacing with data across industries. Recent applications of LLMs to proteins have focused on unsurpervised learning for uncovering and predicting protein structure. This recent paper describes ProteinGPT, a prototype GPT model specialized in learning and understanding protein structures for information retrieval tasks. Utilizing ProteinGPT, researchers can expedite their investigations into protein functionalities and structures.In this recent paper, researchers describe the development of ProteinChat, an LLM-enabled interface that allows users to upload protein 3D structures (e.g., PDB format, widely used for files containing atomic coordinates) and pose questions about the structures and discern insights.
The ProteinChat system consists of three main components:
a GVP-Transformer block
a projection layer
an LLM
The GVP-Transformer block takes the 3D structure of a protein as input and learns a representation that captures important features of the structure. The projection layer further processes the protein embeddings generated by the GVP-Transformer block to align them with the requirements of the LLM. The LLM then uses the encoded protein representation and user prompts to generate relevant answers.
ProteinChat allows seamless interactions and accurate responses based on the protein's structure and user queries. The system uses the Vicuna-13b LLM and a pre-trained GVP-Transformer from ESM-IF1 as the protein encoder. During training, the protein encoder block and the LLM are frozen, and only the projection layer is trained. The RCSB-PDB Protein Description Dataset, constructed from the publicly available RCSB-PDB database, is used to train the ProteinChat model.
Code available here and join the discourse on Twitter here.
Human disease models in drug development [Loewa et al., Nature Reviews Bioengineering, 2023]
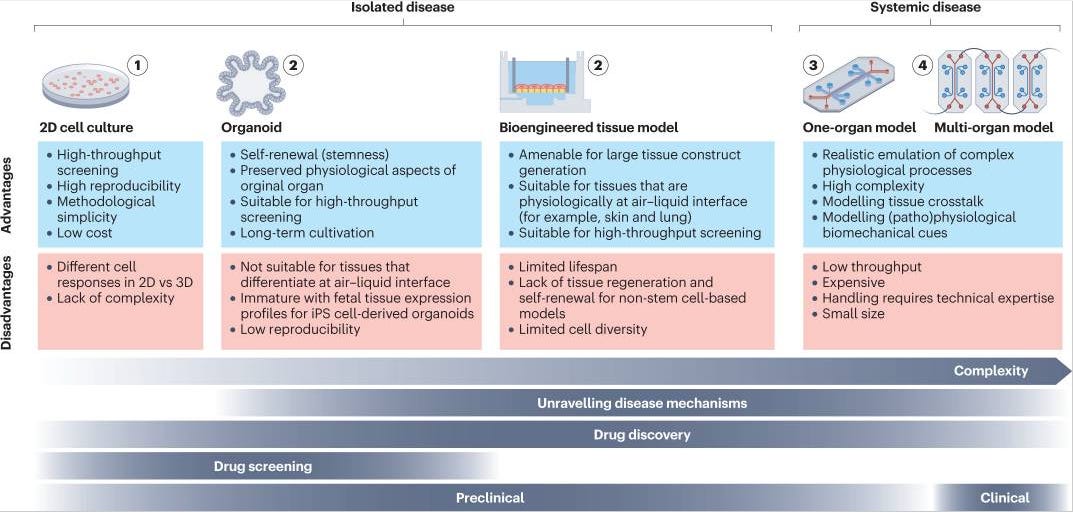
Why it matters: Preclinical models are important for testing experiments in a quick and accurate way; however, accuracy is often lacking given the variability within preclinical models and the clinical setting. New bioengineered human disease models like organ on a chip can accelerate drug development by exhibiting a higher level of clinical mimicry. This review walks through several human disease models that leverage recent advances in bioengineering. A major concern with such preclinical models has long surrounded whether we can model human disease and its various intricacies in a lab-grown tissue or cell culture flask. Yet, with the shortage of non-human primates and lack of clinical predictability, there is a need to innovate new bioengineered preclinical models. The authors highlight three major categories:
Cell culture: 2-D cell culture is often the cheapest, most accessible model for generating preliminary experimental results. The lines can be derived from mouse or human populations and some experiments use engineered reporter lines for manufacturing. As easy as they are to use, 2-D cell culture has its fair share of limitations including unknown translatability and inability to recapitulate physiology. 3-D cell culture addresses some of these issues with more biomimicry but often fall short.
Bioengineered tissue models: generated from human stem cells, these models are 3D printed into a scaffold. They are great at mimicking human tissue and can be used to study layered tissue like the gut epithelia, but are challenged by longevity.
Organoids: these are self-organizing 3D models from derived cells, often stem cells. Various factors influence the development of the cells and allow the tissue to differentiate and regenerate itself. New organoid models are popping up more frequently given new ways to culture cells and change media. Organ on a chip, microfluidic platforms that have mini-tissues, are an extension of organoids (though components are predefined as opposed to self-organized) to study physiology and biophysical and chemical cues.
There are still significant shortcomings marking each of these categories of preclinical models though the need for them is undeniable. The authors walk through some regulatory and clinical considerations for each in this quick, digestible review.
What we listened to
Notable Deals
Roger Perlmutter builds Eikon's pipeline with deal-making flurry, raising $106M more
Italian AAV biotech closes $65M Series A
In case you missed it
Inside the nascent industry of AI-designed drugs
Large Language Models in Molecular Biology
What we liked on Twitter
Events
Field Trip
Did we miss anything? Would you like to contribute to Decoding Bio by writing a guest post? Drop us a note here or chat with us on Twitter: @ameekapadia @ketanyerneni @morgancheatham @pablolubroth @patricksmalone