BioByte 034: advances in prenatal gene therapy, the scoop on Laronde, the three waves of AI in bio, why gene therapies should go virus free, cell atlas of the human lungs
Welcome to Decoding Bio, a writing collective focused on the latest scientific advancements, news, and people building at the intersection of tech x bio. If you’d like to connect or collaborate, please shoot us a note here or chat with us on Twitter: @ameekapadia @ketanyerneni @morgancheatham @pablolubroth @patricksmalone. Happy decoding!
Two posts from us this week! If you only have a minute, here are the highlights:
Gene therapy in the womb is inching closer to reality but delivery remains a challenge
After raising a $440M Series B, Laronde fails to replicate key data in a story reminscent of biotech oversight
AI in Bio take three forms: in silico drug discovery, computer vision and phenotypic drug discovery, and generative AI predictions
The case for why gene therapies should go virus free (hint: has to do with immunogenicity and manufacturing costs)
New bio drop: cell atlas of the human lungs
Deep learning can help us find new antibiotics
we can look to macrophages to help us understand the enteric (gut) nervous system
What we read
Blogs
Gene Therapy in the Womb is Inching Closer to Reality [Levy, Wired, May 2023]
Advancements in in utero gene therapy were presented at the recent American Society of Gene & Cell Therapy meeting in Los Angeles, offering potential solutions for treating genetic diseases before birth. Researchers are exploring the benefits of correcting DNA abnormalities in fetuses, aiming to prevent diseases from developing. Conditions like sickle cell anemia and spinal muscular atrophy, which can be detected during prenatal screening, could benefit from early intervention.
By leveraging the fetus's unspecialized stem cells and underdeveloped immune system, gene therapies have a higher chance of success. However, delivering the therapies to specific organs remains a challenge. Scientists are experimenting with various methods, including optimizing delivery vehicles and timing injections during pregnancy.
One example highlighted in the article is the research conducted on treating spinal muscular atrophy, a genetic disease, using in utero gene therapy in mice. The study demonstrated that the therapy continued to be effective in the fetuses even when the mother had immunity to the editing machinery used. However, in cases where pregnant mice had immunity to the virus used for delivering the gene therapy, more fetal offspring died due to the maternal immune response. This finding suggests that directly injecting the therapy into the umbilical cord early in pregnancy may protect the fetus from the mother's immune response, providing a potential solution for future human trials.
The studies also emphasize the importance of monitoring the reach and effects of genetic treatments. Safety is a paramount concern, both for ensuring proper organ targeting and safeguarding the health of both the fetus and the mother. While progress has been made in animal models, human trials are still several years away. Researchers caution against false hope but remain optimistic about the potential of in utero gene therapy.
The inside story of how data integrity issues roiled Laronde, seen as ‘Moderna 2.0’ [Allison DeAngelis, STAT News, June 2023]
One of the buzziest topics in biotech this week is the story of how Laronde, the Flagship Pioneering biotech that raised nearly $500M, failed to reproduce key data that underpinned their raise. Laronde is focused on circular RNA, a looped RNA molecule that is thought to be more stable and less immunogenic than its mRNA counterpart. Though a couple groups are working on this circular RNA modality, Laronde claimed to have some pretty strong initial results in preclinical models for a GLP-1 drug (you’ve heard of GLP-1 agonists from the weight-loss drugs) and it was really exciting. Months later, no one else was able to replicate those results—a far cry from their public goal to create 100 new therapies in ten years. What’s more—there were some obvious errors in the presented data that suggested it was too good to be true, things like control levels of expression being way off yet no one at the company was able to convince management to take a second look. There were many other warning signs that people, retrospectively, can point to that were left in the shadows before. Allison does an incredible job uncovering and reporting on them in this article.
The main takeaway is that this is unfortunately not an unfamiliar story in biotech. We’ve heard countless stories of promising results with lack of reproducibility, management culture that doesn’t acknowledge all voices, and stone-walling around company milestones. All of these should be warning signs that something is fundamentally wrong. Science isn’t straightforward and lack of reproducibility in results is common. Rather than taking one experiment that happened to go well and running with it, our first instinct should be skepticism. There’s a reason most scientists are trained to not tell anyone about their work until they can prove it at least three times. If only one person can get something to work, that’s not reproducible science. Laronde has since lost several employees and done a round of layoffs, though it doesn’t seem like cash is tight. It will be interesting to see how they emerge from this as circular RNA may still have a lot of untapped potential.
The 3 Waves of AI in Bio [Dr. Omri Drory, NFX, June 2023]
The article discusses the impact of AI in the field of “techbio,” comparing the importance of using AI in bio to the significance of using cloud technology in the tech industry.
The NFX team characterizes three waves of AI in bio:
Wave 1: In-Silico Drug Discovery
The first wave focused on in-silico drug discovery, where AI played a crucial role in scaling up the compound screening process. Companies like Atomwise leveraged AI to train their own Convolutional Neural Networks on vast amounts of biological data, enabling the computational testing of millions of compounds on specific protein targets. While this approach was groundbreaking, it didn't address the challenges of advancing candidates through clinical trials.
Wave 2: Machine Vision + Phenotypic Drug Discovery
In the second wave, AI unlocked phenotypic drug discovery by utilizing machine learning and machine vision. Computers became observational tools, identifying which compounds effectively reversed diseases. This approach, based on Convolutional Neural Networks, allowed for faster experimentation and pattern recognition that humans couldn't achieve alone. Companies like Spring Discovery and Recursion Pharmaceuticals made significant progress in this area.
Wave 3: Generative AI + Protein Structure Simulations
Now, we find ourselves riding the third wave: the combination of generative AI with protein structure simulation. This wave is where things get truly exciting. DeepMind's Alpha Fold amazed the scientific community by accurately predicting the structure of nearly all known proteins. Large Language Models (LLMs), such as those trained on DNA sequences, can now predict amino acids with remarkable accuracy. These advancements have opened the door to designing proteins from scratch. Companies like Zip Therapeutics and Nabla Bio are at the forefront of this wave, generating entirely new proteins with unique functions.
However, it's important to note that relying solely on AI won't make a techbio company stand out for long. It's crucial to have something defensible and patentable. Can the generated outputs actually work? Can they be proven? Answering these questions is essential for success. Additionally, avoiding the middle ground and either being an early adopter or blending multiple waves of technology is key to staying ahead.
The article emphasizes that winning on the business side is equally important. Understanding the market and effectively selling unique offerings are crucial skills. Building partnerships and knowing the target customers are essential for growth.
Intellia’s second CRISPR therapy reduces swelling attacks by 95% in hereditary angioedema [Ryan Cross, Endpoints, June 2023]
Intellia’s NTLA-2002 CRISPR/Cas9-LNP therapy for hereditary angioedema, a rare genetic disease that “leaves people prone to sudden inflammation and swelling” has shown very promising Phase I results. The therapy lowered levels of the inflammation-promoting enzyme kallikrein by inactivating the KLKB1 gene, reducing bouts of harmful swelling by 95%.
The patients, before the therapy, had swelling attacks between once a month to 17 times a month. By the end of the four-month observation period, the patients stopped having attacks and didn’t have one through the 2-10 months later. The therapy demonstrated no severe adverse side effects.
Why gene therapies must go virus-free [Cormac Sheridan, Nature Biotechnology News, June 2023]
Viral vectors for gene therapies like AAVs have important shortcomings that may limit clinical use, such as high manufacturing costs, limited packaging capacity (4.7kb proteins), immunogenicity which limits redosing, and poor tissue specificity. There are several emerging alternative modalities, including engineered lipid or protein nanoparticles, and DNA-based nanocarriers. For example, Aera Therapeutics is developing a protein-nanoparticle system based on endogenous human proteins that assemble into capsid-like structures for the delivery of nucleic acid cargo. A key challenge in the field is tissue selective delivery and liver detargetting, which can act as a “sink” for gene therapy. There are many strategies, such as rationally designing the lipid composition of lipid nanoparticle vectors to selectively bind specific receptors, or building vectors without any inherent tropism for the liver, such as from phage proteins (Gensaic) or DNA-based material (Code Bio). Finally, as transgene expression declines over time resulting in a potential need for gene therapy redosing, non-viral delivery vectors that demonstrate non-immunogenicity will be critical.
Academic papers
An integrated cell atlas of the lung in health and disease [Sikkema et al., Nature Medicine, June 2023]
Why it Matters: This new cell atlas of the human lung provides a centralized, single-cellr reference of unprecedented size. As a publicly available resource, the atlas serves as a central platform for scientists and doctors interested in studying lung biology and developing further research in lung health and disease. Additionally, this project represents a significant milestone towards the larger goal of creating a full Human Cell Atlas, which aims to revolutionize our understanding of biology and disease.Scientists at Helmholtz Munich and their collaborators have created the Human Lung Cell Atlas using artificial intelligence (AI) techniques, mapping the human lung on a single-cell level. The atlas provides a comprehensive understanding of the diversity of lung cell types and their functions in both health and disease. By combining 49 different datasets, including data from over 100 healthy individuals, the researchers were able to analyze how lung cells vary with age, sex, and smoking history. The atlas also includes datasets from various lung diseases, including profibrotic monocyte-derived macrophages in COVID-19, pulmonary fibrosis and lung carcinoma, allowing researchers to compare healthy and diseased lungs and identify shared cell states, potentially leading to new treatment targets and biomarkers for lung diseases.
The creation of the Human Lung Cell Atlas involved integrating data from numerous studies and standardizing cell type annotations to establish a consensus in the field of lung research. With over 2.4 million cells analyzed in the atlas, researchers can identify gene activity in each cell, gaining insights into cell functionality.
Deep learning-guided discovery of an antibiotic targeting Acinetobacter baumannii [Liu et al., Nature Chemical Biology, 2023]
Antimicrobial resistance (AMR) is amongst the most important issues in healthcare. In the US alone, 35,000 people die every year due to antimicrobial resistant microbes, and globally more than 5 million per year. Outnumbering HIV, malaria and tuberculosis combined.
Misaligned incentives mean that pharmaceutical companies don’t develop new antimicrobials. Physicians use new antimicrobial drugs sparingly to delay resistance as long as possible. However, this is in direct opposition to the volume-based business model of the pharmaceutical companies. Given the time and expense required to bring a new antibiotic to market, pharmaceutical companies would rather develop drugs that will bring a higher return on investment. The government has stepped in order to finance the discovery and development of these critical medicines.
In addition, the discovery of new narrow-spectrum, novel chemotype antibiotics that work through new mechanisms of action has been a challenge. New chemotypes, rather than structural analogues, bring long-term efficacy due to the lower prevalence of existing resistance determinants.
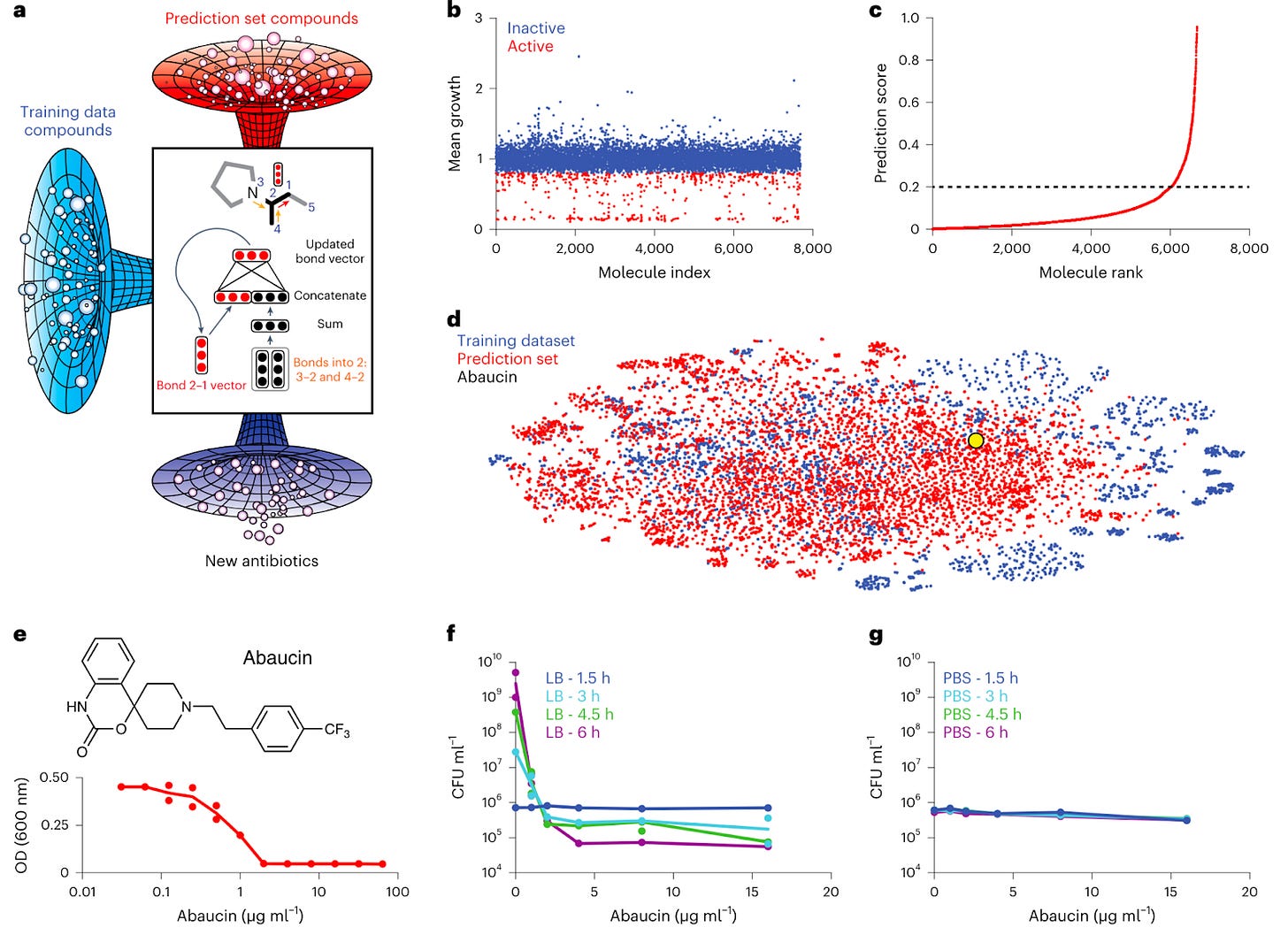
Liu et al. developed a deep learning methodology to identify narrow-spectrum, novel chemotype antibiotics. The authors identified abaucin, an antibiotic against Acinetobacter baumannii, a bacteria that “displays multidrug resistance due to its robust outer membrane and its ability to acquire and retain extracellular DNA that frequently encodes antibiotic resistance genes”.
In order to the find the company, the authors:
Screened a selection of > 7,500 small molecules for those that inhibited the growth of A. baumannii in vitro
This data set was used to train a binary classifier to predict whether structurally new molecules may display activity against A. baumannii by leveraging a directed message-passing neural network architecture, which translates the graph structure of a molecule into a continuous vector. The final vector is then supplemented with RDKit and fed into feed-forward neural network that predicts antibacterial properties.
The models were then applied to a new set of molecules, returning a prediction score for each compound, representing the probability of growth inhibition against the bacteria
Through stringent prediction score cut-offs and molecule structure assessment, the authors found two compounds as potential candidates, one of which was aubacin.
Large Scale Foundation Model on Single-cell Transcriptomics [Hao et al., BioRxiv, June 2023]
Regular readers of the newsletter are well aware of our interest in foundation models in bio. This week, another preprint was published building pre-trained, transformer-based models to transcriptomic data. A few highlights from the paper, and what’s novel relative to previously published papers covered in the newsletter:
The FM was trained on one of the largest single-cell datasets to date (50M human single-cell transcriptomes, resulting in a 100M parameter model).
Perhaps the most important innovation of the paper was the pre-training task. The authors developed a method called read-depth-aware (RDA) modeling, which incorporated read depth (the number of times a transcript is sequenced, which impacts the quality of the data) into the training task. The pre-training task was a masked gene prediction task, but the model used a low-read depth variant of cell gene expression to predict the masked gene. This allowed the model to learn gene-gene relationships that were robust to differences in read depth.
The FM was fine-tuned on downstream tasks including gene expression enhancement, tissue drug response prediction, single-cell drug response classification, and single-cell perturbation prediction. Most interesting/novel was drug response prediction. Embeddings (lower dimensional representations of high-dimensional input data) generated by the FM significantly improved the prediction of drug IC50 across a range of chemotherapies and targeted therapies.
Dedicated macrophages organize and maintain the enteric nervous system [Nature, June 2023]
Why it matters: The enteric nervous system (ENS) – the neuronal system responsible for gut function – independently controls necessary functions, such as intestinal peristalsis, secretions, and blood flow. At birth, the ENS is notably immature and requires significant processing in order to be functional in adulthood. Here, the authors demonstrate that resident macrophages of the muscularis externa (MMϕ) prune synapses and phagocytose neurons in early development, and over time, develop a neurosupportive phenotype akin to glial cells in the CNS. Depletion and/or dysfunction of these macrophages may underlie a new mechanism of action of gut-oriented disease pathophysiology. The authors first demonstrate that depletion of MMϕ during a certain refinement period in development led to hyperganglionosis and deranged ENS structure, demonstrating their role in reorganizing the ENS during early development; in contrast, depletion at a later point did not influence neuronal density, suggesting a critical period of refinement. Furthermore, early depletion of MMϕ led to faster intestinal transit in adulthood. Furthermore, in adulthood, MMϕ demonstrate a neurosupportive function; scRNA-seq identified a unique population physically associated with neuronal cell bodies and fibers (NA-MMϕ); this population may represent a unique niche that is critical for neuronal survival in adulthood. Indeed, loss of MMϕ led to loss of enteric neurons and impacted transit time.
Additional studies demonstrated that these MMϕ were almost analogous to microglia in the brain; TGFβ release by the ENS was identified to influence local macrophages (just as for microglial programming in the brain), and led to MMϕ exhibiting an upregulation of genes identified in NA-MMϕ. Furthermore, MMϕ demonstrate significant transcriptional and functional plasticity (again, similar to microglia), with upregulation of phagocytic and replication genes during early life, and later in adulthood, immune surveillance programs.
Although several questions still remain, this work sheds light on important dynamics underlying the ENS-immune axis, and opens up a new avenue of investigation for understanding intestinal disease.
What we listened to
Notable Deals
Novartis buys Chinook for up to $3.5B to expand kidney disease pipeline
Beacon launches with $120M to develop eye gene therapies
Hopewell Tx raises $25M for delivery
Bitterroot Bio raises $145M to take cancer target and expand to heart medicines
In case you missed it
Decoding Biosecurity & Biodefense: Building A Global Immune System
Alex Telford published a list of recommended resources for keeping up with the biotech industry. We were honored to be included! Find the full list here.
What we liked on Twitter
Resources for public genomics data @tangming2005
Fundraising tips for biotech founders @angelosgeo
State of the biotech pipeline @maxjacobsCFA
Market deep dive on digital pathology @davidwest_irl
Pseudocode and what that means for defensibility
Field Trip
Did we miss anything? Would you like to contribute to Decoding Bio by writing a guest post? Drop us a note here or chat with us on Twitter: @ameekapadia @ketanyerneni @morgancheatham @pablolubroth @patricksmalone