BioByte 036: the future of single cell bio, cell and gene therapy manufacturing, knowledge graphs for biomedical data, secret messaging with endogenous chemistry
Welcome to Decoding Bio, a writing collective focused on the latest scientific advancements, news, and people building at the intersection of tech x bio. If you’d like to connect or collaborate, please shoot us a note here or chat with us on Twitter: @ameekapadia @ketanyerneni @morgancheatham @pablolubroth @patricksmalone. Happy decoding!
Short on time? Here’s what we covered this week:
A primer on the current state and future of single-cell biology
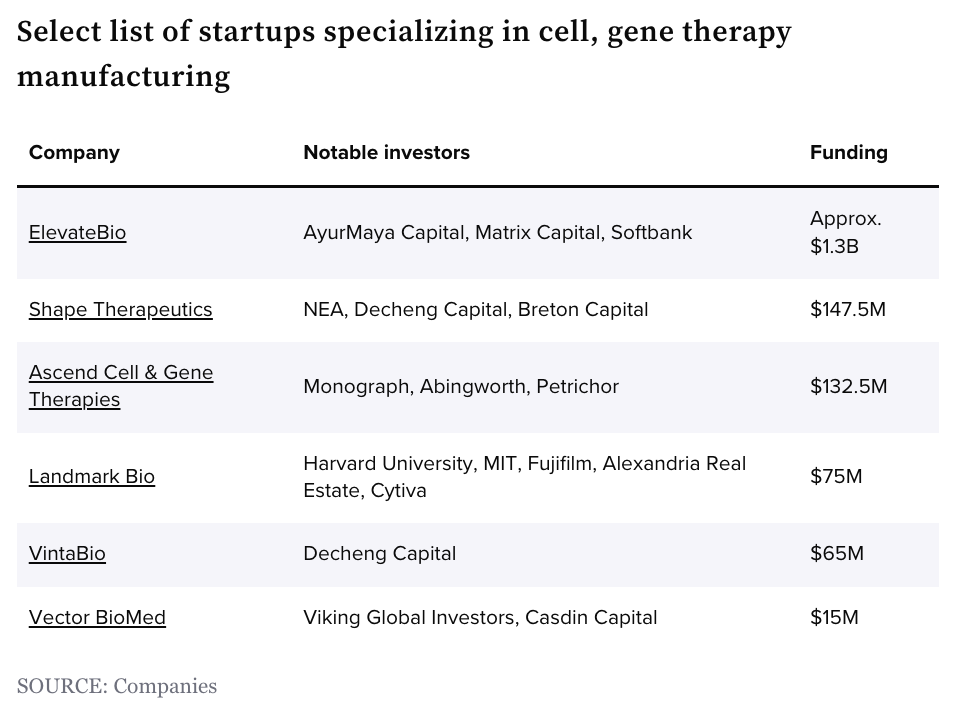
Manufacturing for cell and gene therapies: current challenges and opportunities
Sarepta became the first company to have an approved gene therapy for Duchenne muscular dystrophy on the market
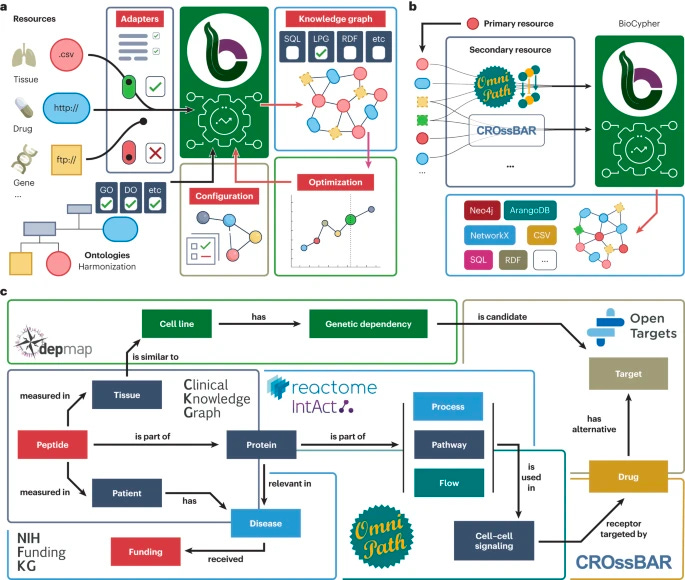
BioCypher offers a technical framework for creating reusable and accessible knowledge graphs for biomedical representations
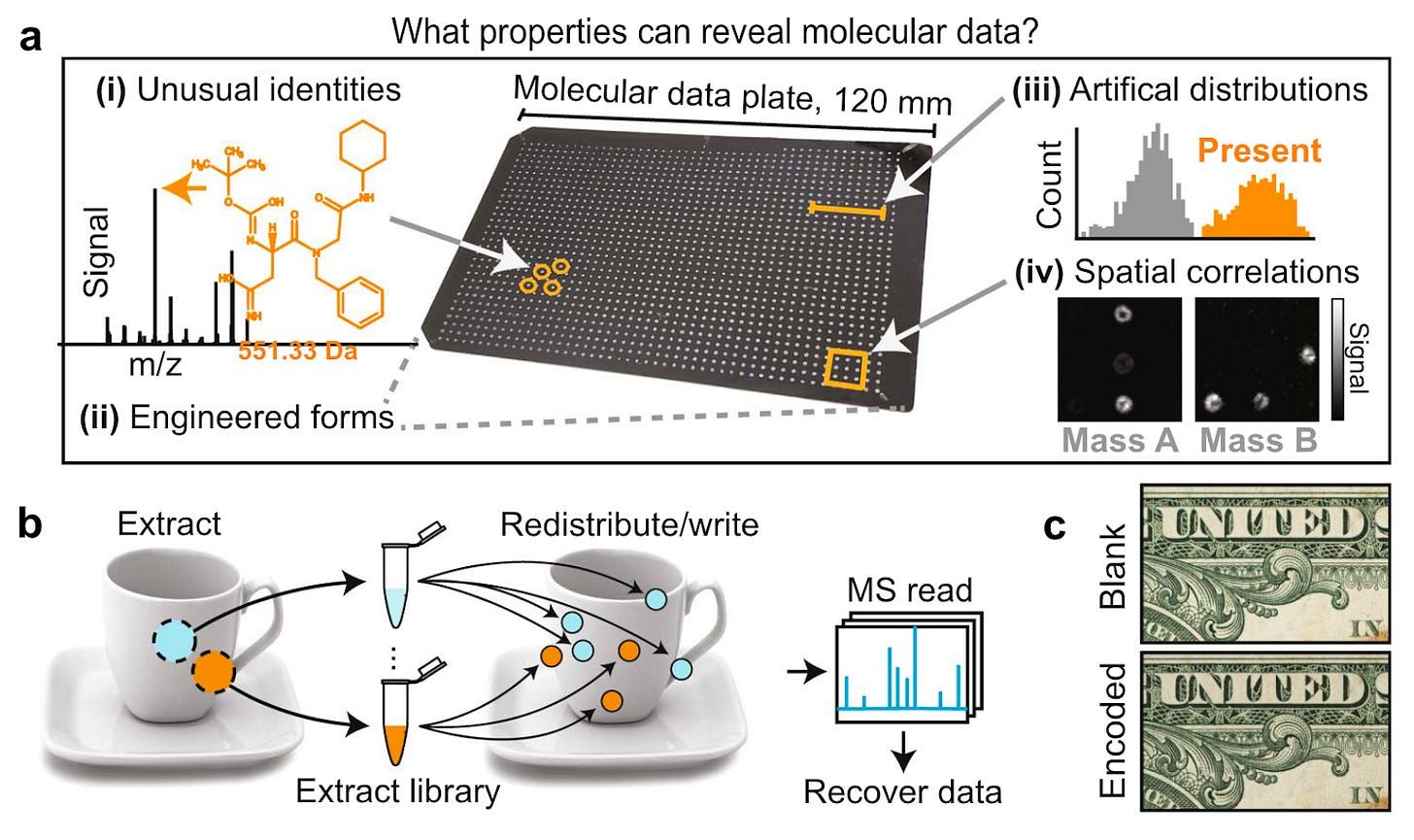
Researchers are experimenting with data encoded in molecular domains and are demonstrating the potential of chemical fingerprints for secret messaging and extreme information density
What we read
Blogs
Single-cell biology: what does the future hold? [Polychronidou et al., Molecular Systems Biology, June 2023]
Single cell biology has exploded over the last several years. Unlike analysis of bulk tissue samples, single cell analyses can resolve cellular heterogeneity, revealing specific cell types and cell states in disease contexts. In this editorial in Molecular Systems Biology, recent developments, challenges, and opportunities in the single cell field are discussed.
Some highlights:
The application of deep learning, and in particular LLMs, to large single-cell datasets will aid in the standardization of disparate and heterogeneous datasets.
Physical interactions between cells might be as important as molecular signals for determining cell fate and behavior. integrating in vitro model systems like organoids that permit mechanical control of the 3D environment will be important for this.
Improved understanding of the principles of tissue self-organization to inform methods for integrating measurements that cross biological scales (eg transcriptomics, proteomics, metabolomics, cell morphology)
The incorporation of a temporal dimension into single-cell biology, moving from spatial biology to temporal biology.
Single cell proteomics will be critical in determining the correlation (or lack thereof) between mRNA expression changes and protein concentration. mRNA concentrations in a cell are highly dynamic and can be transient, and not all mRNA is translated into protein.
Cell and gene therapy manufacturing: the next generation of startups [Gwendolyn Wu, BiopharmaDIVE, June 2023]
Last week we talked about the overall cell and gene therapeutic landscape, and this week we’re continuing the thread with a highlight on manufacturing. Developing cell and gene therapies is a complex and costly endeavor, with manufacturing presenting significant challenges. Key bottlenecks include the intricate process of assembling specialized components and the limited capacity of large contract manufacturers.
Specialized Components: Cell and gene therapies require the use of materials not commonly used in other pharmaceutical products. The construction of these therapies involves synthetic genetic material, vectors (benign viruses), and specialized lipid bubbles, making the manufacturing process intricate and challenging, especially on a large scale.
Limited Capacity and Long Waitlists: Large contract manufacturers often have limited capacity, leading to long waitlists for startups and academic labs. This delay can hinder progress, causing missed milestones and endangering future funding. Unlike established biotechs, startups cannot afford to build their own manufacturing plants, making it crucial to find alternative solutions.
Technology Transfer and Troubleshooting: Transferring technology from small research labs to larger organizations can be arduous and prone to glitches. Contract development and manufacturing organizations (CDMOs) may prioritize work with larger biotech and pharmaceutical firms, leaving startups with limited options. Troubleshooting and optimizing the manufacturing process for complex therapies require significant oversight and expertise.
Cost and Ownership: Establishing internal manufacturing capabilities can be costly for startups. Viral vectors, for example, are expensive to produce and handle. Owning manufacturing facilities comes with financial risks, as the success of the therapy determines the return on investment. Companies need to balance the need for in-house manufacturing against the potential financial burden.
Pricing Considerations: Cell and gene therapies are among the costliest medicines to produce, and manufacturing improvements could provide an opportunity to reassess pricing strategies. High production costs have led to price tags ranging from hundreds of thousands to millions of dollars for approved therapies. Addressing manufacturing challenges may enable more cost-effective production, potentially influencing the pricing of these therapies.
Ozempic 3.0? Lilly’s ‘triple-G’ drug shows biggest weight loss yet in mid-stage trial [Chen, STAT, 2023]
Retatruride, Lilly’s weight loss drug, has shown a 24.2% weight loss in a mid-stage trial. This is the greatest amount seen yet with an obesity drug. Retatruride targets three hormones (GLP-1, GIP and glucagon), rather than two like Lilly’s Mounjaro (GLP-1 and GIP) or a single hormone-like Ozempic/Wegovy (GLP-1).
The drug was linked to gastrointestinal side effects for which the rates varied depending on dose: nausea ranged from 14-60%, vomiting ranged from 3-26% and one person taking the highest dose experienced acute pancreatitis. There were also instances of arrhythmia, increased heart rate and one patient experienced prolonged QT syndrome. Another concern is on the type of weight loss: the aim is to reduce fat, rather than muscle and this was not measured in the study.
There will be more attention paid about the quality of weight loss now that the weight loss is so large.
FDA grants conditional approval to Sarepta’s gene therapy for Duchenne muscular dystrophy [Mast and Feuerstein, STAT, June 2023]
Sarepta became the first company to have an approved gene therapy for Duchenne muscular dystrophy on the market. Clocking in at $3.2M, it is the second most expensive gene therapy, after Hemegenix at $3.4M, a hemophilia treatment. This dystrophy drug, which works by delivering a version of the disrupted gene dystrophin to cells, will be limited to boys aged 4-5 right now as the one randomized trial did not show broad effect over placebo in all patients. This restriction may change after a larger randomized trial at the end of the year. Interestingly, the dystrophin produced by the therapy is very different from human dystrophin as it is shortened, but does show that we are able to get dystrophin into the human body in the first place.
Engineering Biology: How to Build Data Centric Biotech [Jacob Oppenheim, Digitalis, June 2023]
Jacob takes to the Digitalis blog to write about data science and AI in biotech…starting with the prescient quote “if you don’t have novel and effective science in the first place, no amount of data science will save you.” AI and data science are most successful in science when they are not needed in the first place, and the first roadblock is lack of data. How do you get more data? Better science. Jacob outlines two strategies for solving the lack of data problem that comes up time and time again in this newsletter and every comp bio company.
Better tools and systems for data management. Biotech lacks standardization—both between individuals and between institutions. We need to be consistent and cognizant of metadata and context in information collection. “Digitized biotech” scales faster.
Building around high throughput biology—this is an interesting point and one that isn’t as obvious but easier to put into practice. Building around high throughput biology involves reconstructing drug development around a process that a lab or company does really well. Using combinatorial chemistry to find small molecules. Using microfluidics to develop antibodies. Even without computation, theoretically, these experiments would deliver interesting results.
Academic papers
Democratizing knowledge representation with BioCypher [Lobentanzer et al., Nature Biotechnology, June 2023]
Why it matters: The rapid growth of biomedical data and the increasing importance of machine learning tools have highlighted the need for efficient knowledge representation. Knowledge graphs (KGs) have emerged as a popular form of knowledge representation, but building task-specific KGs can be costly and challenging. In response, the BioCypher framework has been developed to support the creation of KGs, addressing issues of modularity, harmonization, reproducibility, reusability, and accessibility. By providing a low-code access point and leveraging existing resources, BioCypher simplifies the KG building process, facilitating interoperability and enhancing research efficiency in the biomedical community.With the exponential growth of biomedical data, knowledge graphs have become a favored method for representing complex information in a graph format. However, creating task-specific KGs remains a challenge, as different research areas require distinct structures and content. The BioCypher framework addresses this issue by enabling the development of customized KGs tailored to specific research tasks.
The current landscape of biomedical KGs lacks adherence to FAIR and TRUST principles, impeding navigation and integration. Many KGs are built manually for specific applications, leading to a lack of consistency and hindering comparison and combination efforts. BioCypher addresses these challenges by emphasizing modularity, harmonization through ontologies, and enabling reproducibility and subgraph extraction, resulting in more reusable and accessible KGs.
BioCypher is implemented as a Python library that offers a low-code approach to data processing and ontology manipulation. The framework embraces a modular architecture, allowing the reuse of data and code through input, ontology, and output adapters. It provides access to ontologies for harmonization, facilitates reproducibility of KG structures, and ensures reusability and accessibility through open-source development and adherence to FAIR and TRUST principles.
The platform's modular nature offers benefits such as reduced developer time and improved decision-making on knowledge representation. It supports various output adapters for database management systems, including popular graph and relational databases, enhancing performance optimization. Application programming interfaces (APIs) built on top of BioCypher KGs enable versatile queries, user interaction, and integration with machine learning frameworks, natural language processing applications, and standard pipelines.
Secret messaging with endogenous chemistry [Kennedy et al., Nature Scientific Reports, 2021]
Why it matters: “Secrecy systems ought to be designed under the assumption that an enemy can apply unlimited resources to intercept a message, so if molecular steganography is used for security, what properties of chemically embedded covers might alert adversaries or eavesdroppers to the presence of a message?”In this paper published in 2021, the authors describe common vulnerabilities in molecular data systems:
In DNA storage, for instance, the presence of PCR primers, terminus tags and heavily-amplified oligomers are all detection risks that could indicate the presence of digital data.
In chemical datasets, the presence of unusual chemical structures could be used to discriminate between embedded covers from other objects. Even when using common molecules, the authors state that other noticeable features such as correlated concentration profiles, atypical isotope ratios or bimodal concentration distributions can risk exposing communication to third parties.
To overcome these vulnerabilities in molecular stenography, the authors used existing chemistry in the original substrate of the object and a concentration redistribution through a non-random spatial pattern to encode data. In their case on a one dollar bill.
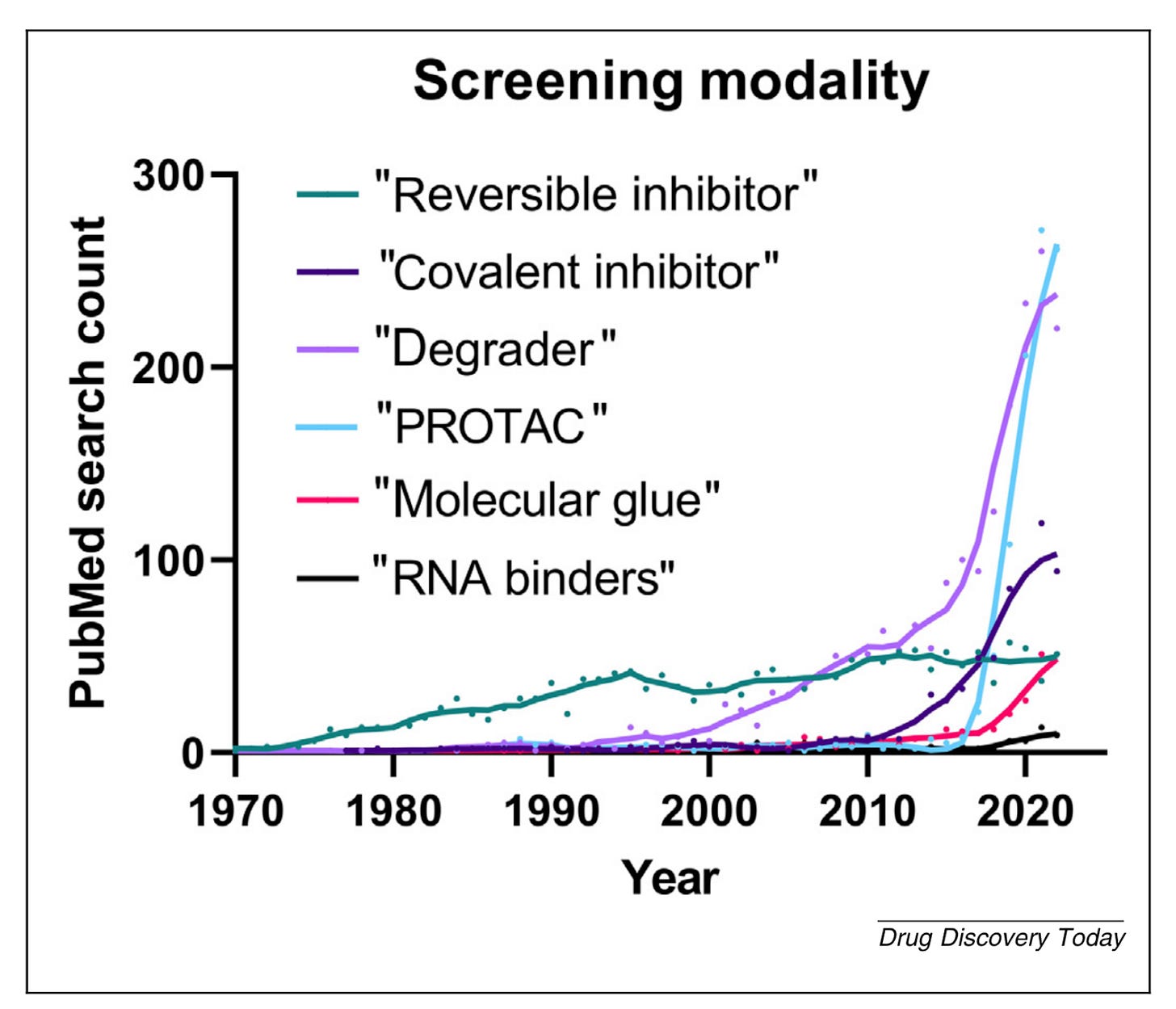
A perspective on the changing landscape of HTS [Lanne et al., Drug Discovery Today, July 2023]
Why it matters: As biopharma increasingly targets historically intractable target classes such as transcription factors and RNA, the screening methods used to generate therapeutic agents are also changing. Newer screening methods are focused on characterizing complex molecular interactions in addition to activity-based readouts. There will be no one method to rule them all, however, and the choice of screening method must be tailored to the specific therapeutic modality and target class. Target-based drug discovery begins with a therapeutic target (usually a protein), followed by screening methods to discover or design a therapeutic agent (eg a small molecule or a biologic) that modulates that target's activity. Many approved drugs today bind to "tractable" targets that contain well-defined binding pockets, however, the prevalence of these targets in AstraZeneca's portfolio has decreased over time. In this review, the AstraZeneca Hit Discovery team reviews a recent shift to biologically-validated but difficult-to-drug targets, including new chemical modalities for modulating historically undruggable targets.
The most tractable/druggable drug targets tend to be kinases, GPCRs, and protein transporters. Less druggable classes without well-defined binding pockets include transcription factors, RNA, and scaffold proteins. Transcription factors, for example, are difficult to drug because ~80-90% of TFs exhibit a high degree of intrinsic structural disorder (biotechs like Peptone and New Equilibrium Bio are focused on drugging intrinsically disordered proteins).
New small-molecule modalities have emerged to address these challenging target classes (Figure below), including targeted protein degradation like PROTACs and molecular glues and covalent inhibitors. With a greater focus on intractable targets, screening methodology has changed as well. Traditionally, screening methods have consisted of biochemical or cell assays with functional or activity-based readouts. Newer methods, however, have shifted to measuring more complex molecular interactions instead of or in addition to activity-based readouts. A few of the more commonly used approaches:
Affinity selection mass spectrometry, which separates unbound compounds from protein-ligand complexes, allowing bound compounds to be detected via mass spec.
Cellular thermal shift screening assays work on the principle that compound binding alters the thermal stability of a protein. The advantage of this approach for intractable targets is that because the screen occurs in cells, expression and purification is not necessary (a challenging process for many intractable targets).
Protein interaction assays that monitor for targeted protein degradation of a library of chemical matter with the potential to be used in molecular glues or PROTACs.
What we listened to
Notable Deals
MoonLake's mid-stage IL-17 data sent shares skyrocketing. Now it plans to sell $250M in stock
Startup Tagworks raises $65M to make ‘click-to-release’ cancer drugs
Databricks picks up MosaicML, an OpenAI competitor, for $1.3B
In case you missed it
Moderna is 'laser-focused' on AI, from clinical development to marketing
Sanofi is “all in” on artificial intelligence and data science to speed breakthroughs for patients
What we liked on Twitter
How to attract top biotech talent. @angelosgeo
ML strategies to improve performance when data is limited. @AlissaHummer
Will big pharma go on acquisition sprees to replace revenue after loss of exclusivity? @MichaelRetchin
Biological models vs. biological reality. @PatrickMalone
Field Trip
Did we miss anything? Would you like to contribute to Decoding Bio by writing a guest post? Drop us a note here or chat with us on Twitter: @ameekapadia @ketanyerneni @morgancheatham @pablolubroth @patricksmalone