BioByte 041: NME generation: pharma vs. biotech, validation in AI drug discovery, whole body cellular mapping, deep learning for synergistic drug combos, AI x brain-computer interfaces
Welcome to Decoding Bio, a writing collective focused on the latest scientific advancements, news, and people building at the intersection of tech x bio. If you’d like to connect or collaborate, please shoot us a note here or chat with us on Twitter: @ameekapadia @ketanyerneni @morgancheatham @pablolubroth @patricksmalone. Happy decoding!
Hope everyone’s enjoying what’s left of summer. Going away this weekend? We’ve got you covered. As always, here’s a rundown of this week in bio:
Assessing the current state of innovation: pharma vs. biotech and the origins of new molecular entities and therapeutic biologics
A call for better validation benchmarks and datasets in drug discovery
An expert guide on how to sell AI products to pharma
Whole-body cellular mapping pushes our current understanding of histology and spatial biology forward
Deep learning models uncover novel synergistic drug combinations
Curated resources dedicated to AI (ML/DL/RL) applied to brain-computer interfaces
What we read
Blogs
Investigating the origins of recent pharmaceutical innovation [Schuhmacher at al., Nature Reviews Drug Discovery, July 2023]
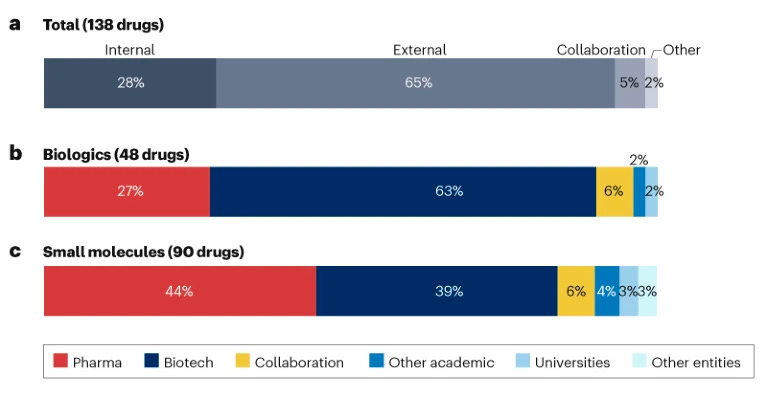
Given the increasing level of collaboration between pharma and biotech companies through M&A and partnerships, we can’t discern where new molecular entities (NMEs) and new therapeutic biologics (NTBs) arise from. The authors systematically assessed the origins of innovation for NMEs and NTBs approved between 2015-2021 by the CDER of the FDA for the top 20 biopharma companies globally.
Of the 323 new drugs approved by the FDA, 138 were filed by top 20 biopharma companies (~43%). The majority (65%) of these originated from external sources, perhaps unsurprisingly, and 28% invented internally and only 5% discovered collaboratively.
The authors define biotech by foundation date (a biotech = founded after 1976). Biotech companies invented about half of the 138 drugs approved, and this is heavily skewed towards biologics. Given the extensive reliance on biotech company innovation, they suggest a new term for them: instead of a FIPCO, we should name them a BIPCO (biotech-leveraged pharma company).
We Need Better Benchmarks for Machine Learning in Drug Discovery [Pat Walters, Practical Cheminformatics, August 2023]
In the field of AI-enabled drug discovery, many papers report a benchmark comparing their algorithm and/or molecular representation with the current state of the art. However, these benchmarks are often not well-curated or relevant to the problem at hand. This makes it difficult to compare different methods and identify the best approach for a particular application.
Pat Walters, Chief Data Officer at Relay Therapeutics, argues that the best way to make progress in the field is to fund a large public effort to generate high-quality data and make it available to the community. This would allow researchers to compare methods on a more level playing field and identify the strengths and weaknesses of different approaches. He argues that this data must be:
Relevant. For example, a benchmark for predicting the toxicity of a molecule should use data that includes molecules that are known to be toxic.
Well-curated. This means that the data should be clean and free of errors.
Current industry standards, such as the MoleculeNet dataset, are not well-curated and may not be representative of the real world. This means that results obtained on this dataset may not be generalizable to other applications.
Walters concludes by stating that we need to move beyond "meaningless benchmarks" and focus on developing methods that are truly useful for drug discovery. This will require us to carefully evaluate benchmark data, identify the strengths and weaknesses of our methods, and develop strategies to overcome the limitations of current methods.
Will unpredictable side effects dim the promise of new Alzheimer’s drugs? [Jennifer Couzin-Frankel, Science News, August 2023]
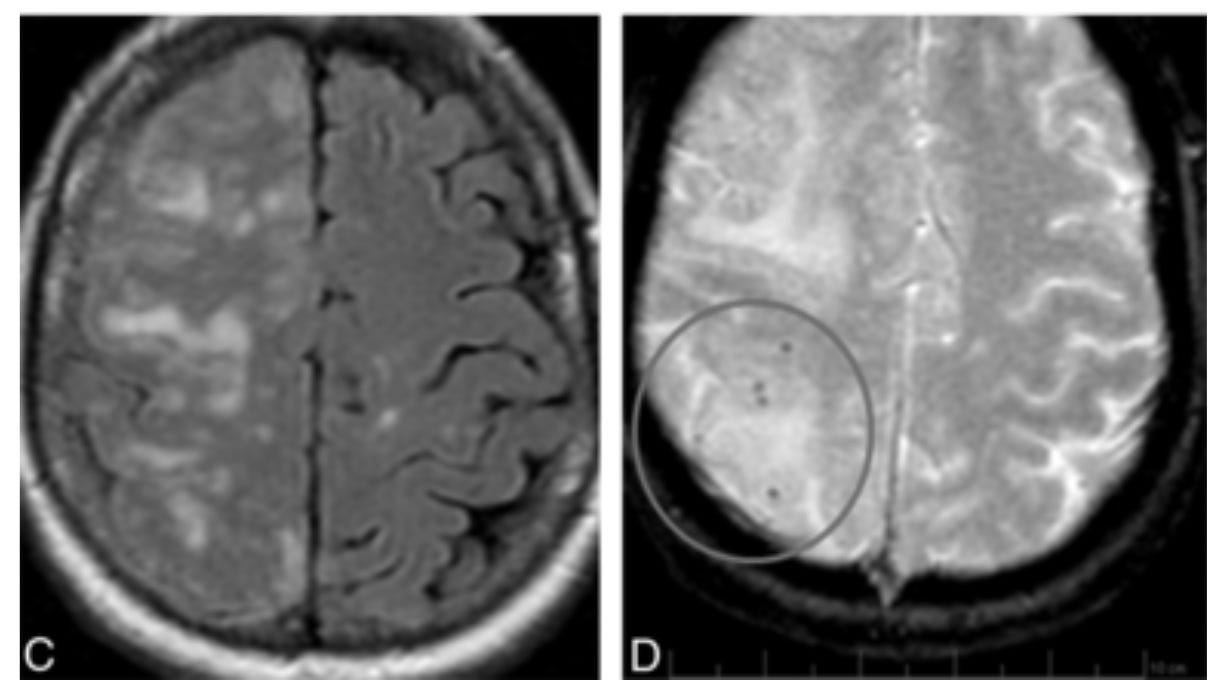
For the first time, a number of disease-modifying therapies for Alzheimer’s are about to hit the market. These drugs are all monoclonal antibodies that bind to and clear amyloid from the brain. Clinical adoption may be limited, however, due to a potential serious side effect called amyloid-related imaging abnormality (ARIA). ARIA causes brain swelling and bleeding, can affect up to ⅓ of patients, and can range in severity from asymptomatic to fatal. The cause of ARIA is unknown, and is under-investigated - there is only one current NINDS grant investigating the cause of ARIA.
The grant’s lead investigator, Donna Wilcock, believes that cerebral amyloid angiopathy (CAA), a buildup of amyloid in brain blood vessels, plays a role in the development of ARIA. The hypothesis is that hemorrhages are caused by the breakdown of amyloid deposited in the blood vessels. Monoclonal antibodies bind to amyloid, recruiting microglia activation and recruitment which cause inflammation, swelling, and potentially hemorrhage. Further research is needed to elucidate 1) the exact MoA of ARIA, 2) if and when to return patients to antibody treatment after developing ARIA, and 3) how to treat ARIA (some evidence that steroids can be effective).
Edema (brain swelling) from ARIA shown on MRI (C) and associated hemorrhage (D).
Selling AI Products + Services to Big Pharma [Brandon White, August 2023]
Brandon White, an experienced engineer and angel investor, published what we have found to be one of the most comprehensive guides on selling AI to pharma, whether it is a tool, service, or platform. He starts off with the three truths of AI in drug discovery, which we will boil into one truth: the closer you are to influencing and marketing drugs themselves, the more value you create. He’s also refreshingly honest—pharma doesn’t care about buzzwords. Language like “AI to develop drugs faster and cheaper” will not land you meetings. More specific language around developing approved drugs will, for example “know the toxicity of your drug before running animal models”. So much more power in one sentence.
The other main point that Brandon touches on is building trust with AI. This can be through transparency on what training data looks like, discussing interpretability of the models, focusing conversation on use-cases as opposed to academic benchmarks, and how AI is tangibly working with data to improve decision making (namely, reducing failure rate). There is a lot of hard-earned knowledge packed in this post making it a must-read for anyone selling to pharma.
Academic papers
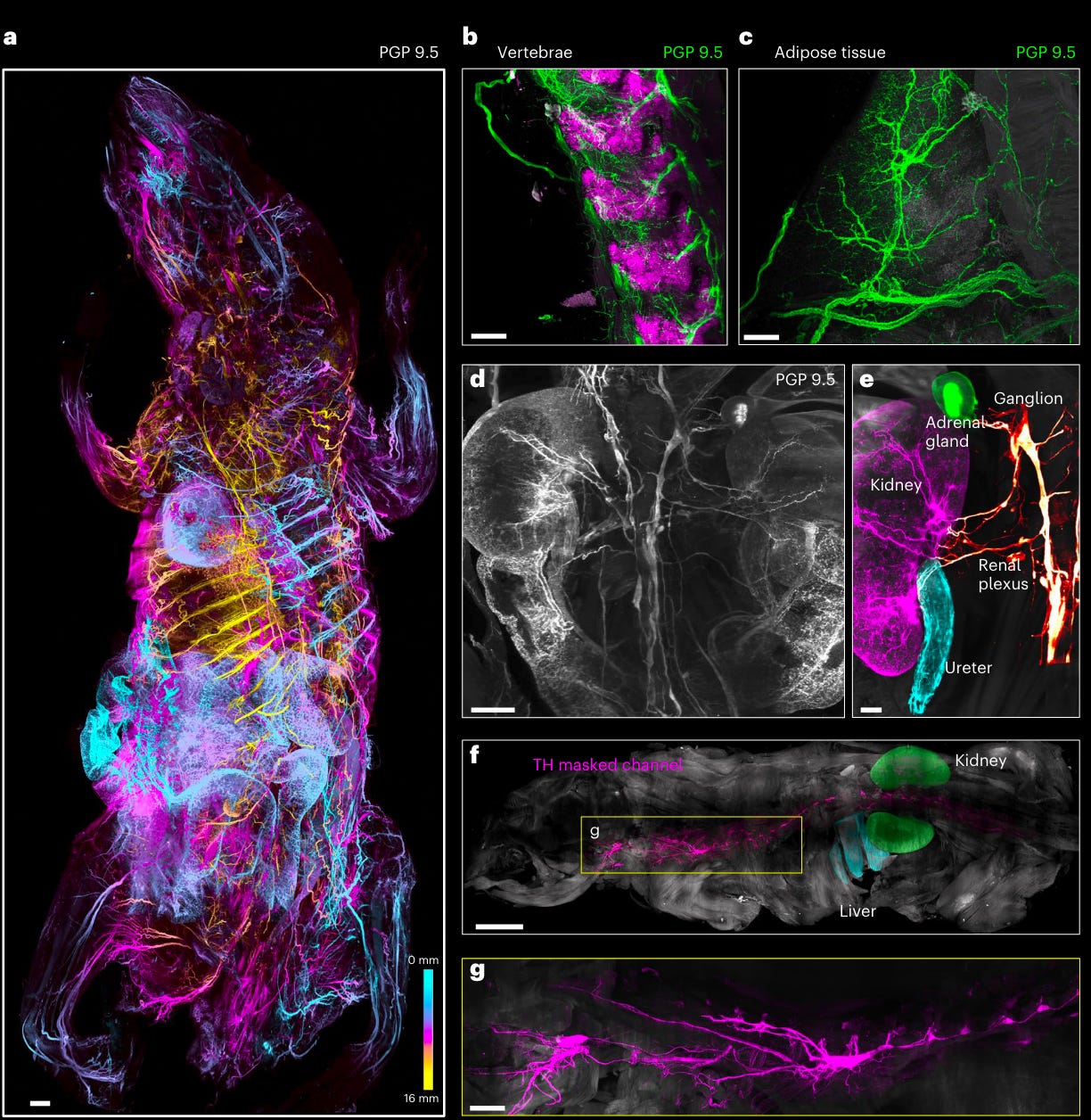
Whole-body cellular mapping in mouse using standard IgG antibodies [Mai et al., Nature Biotechnology, July 2023]
Why it matters: Mapping the distribution, connectivity and molecular makeup of cell types across the whole body is a present challenge in histology and spatial biology. For example, cellular-level maps showing the interconnection between peripheral and central nerves is still lacking. In the context of nerve imaging, most methods rely on transgenic animals which limits experimental design flexibility and can be prohibitively expensive. The ability to have whole-body connectivity maps will be required to understand “functional interdependence” of organ systems in healthy and diseased states.Recent clearing technologies (like DISCO) have enabled labeling and imaging of intact tissues, organs and bodies. However, with other whole-body imaging techniques, that coupled with these clearing technologies, required transgenic expression of fluorescent proteins or the use of nanobodies. Transgenic expression can limit experimental design and there are a limited number of nanobodies that are functional in a histological context.
IgG antibodies, however, are standard in histological labeling yet due to their size lack the permeability granted by the clearing techniques. The authors of this paper developed a new method, wildDISCO, which enhances the penetrance of standard (150kDa in size) antibodies into a whole mouse.
Their method uses “cholesterol extraction for permeabilization to ensure homogeneous penetration and staining across the tissues of the whole mouse body including muscles, bones, the brain and the spinal cord.” The beautiful neuroanatomical and lymphatic mapping can be seen above.
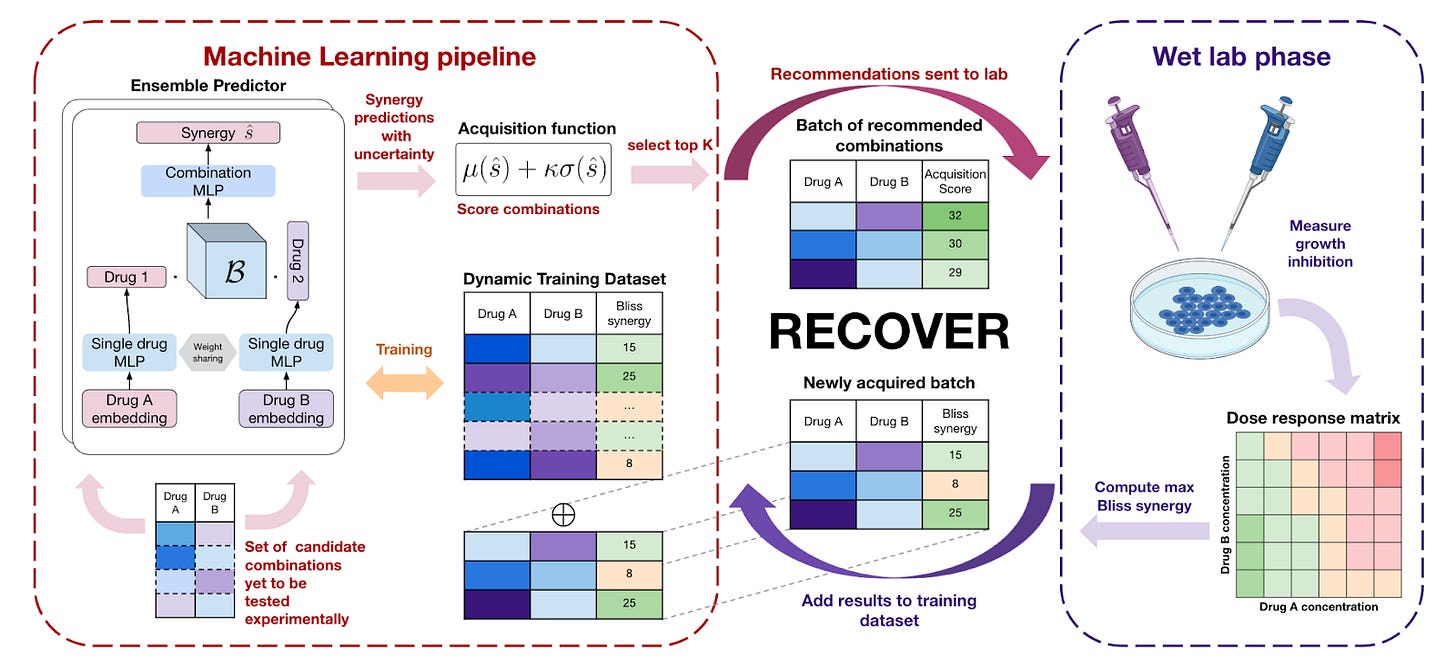
RECOVER: sequential model optimization platform for combination drug repurposing identifies novel synergistic compounds in vitro [Bertin et al., arXiv, 2023]
Why it matters: Drug combinations are important in the treatment of disease that is subject to evolutionary dynamics (i.e. cancers and infections), as cancerous cells and pathogens can evolve to become resistant to a single drug. Discovering synergistic drug combinations can aid patients by improving overall efficacy, whilst potentially limiting adverse effects. However, testing the entire combinatorial space for all possible drugs is not tractable and therefore understanding “where” to look is required to find meaningful synergistic drug combinations with minimal wet lab experimentation.In this paper, the team at Relation Therapeutics and MILA present RECOVER: a deep learning regression model that predicts the synergy score using molecular fingerprints as inputs. RECOVER is a sequential model optimization platform that can guide wet lab experiments to exploit the most relevant areas in combinatorial space by selecting the experiments (pairs of drugs) that it would like to evaluate. After each round of experimental evaluation, the model is iteratively trained with new data, which allows performance to improve (I.e. finding better synergistic pairs of drugs).
Whilst the entire paper is worth a read, briefly, their results demonstrate a 5-10X better estimate for the enrichment of synergistic drug pairs when compared to selecting drug combinations at random and 3X better versus a single batch using a pre-trained model. Importantly, they demonstrated that they could rapidly discover drug combinations with similar mechanisms and efficacy to those already in clinical trials. Thanks Simon B. for the heads up!
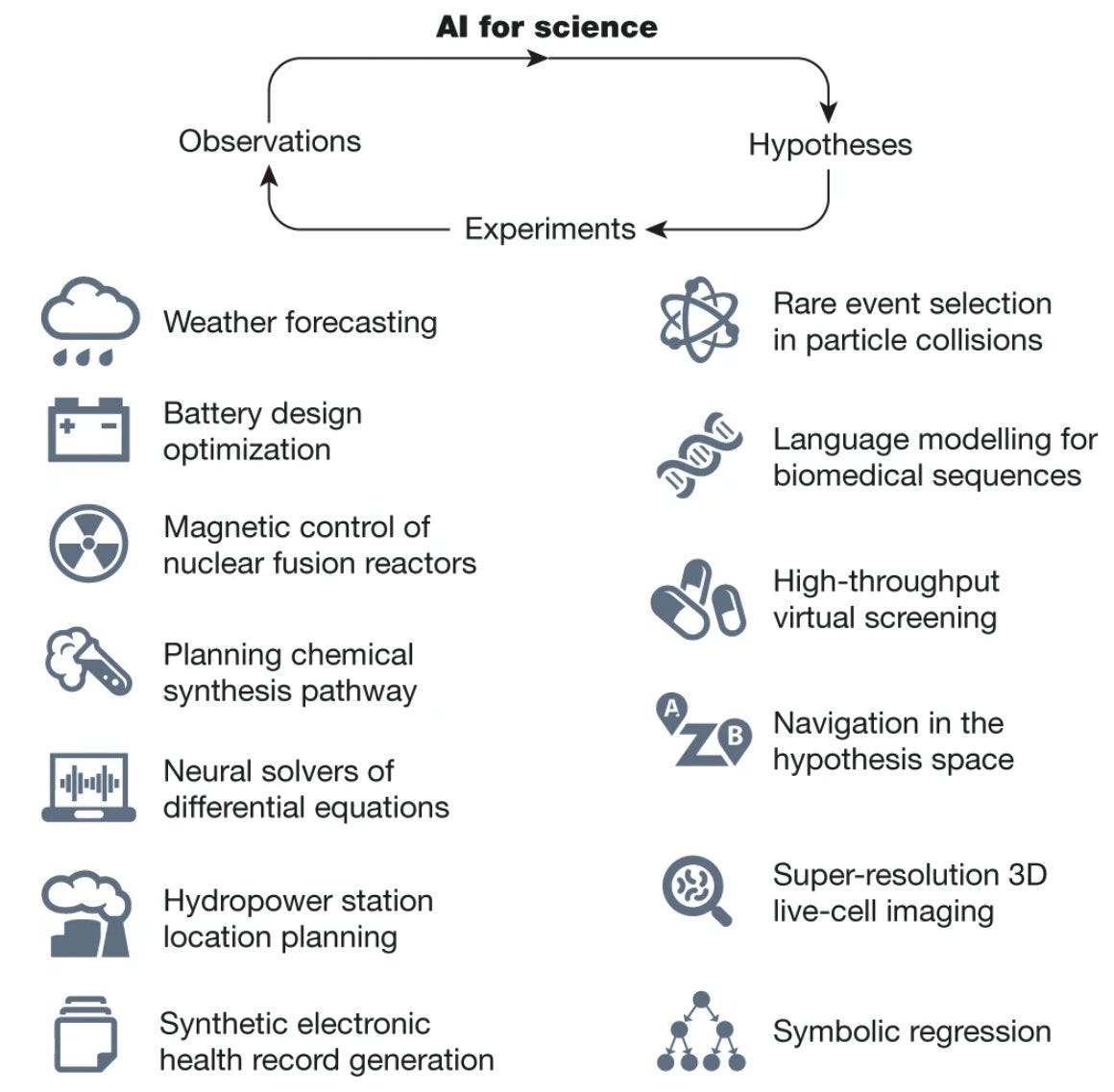
Scientific Discovery in the Age of Artificial Intelligence [Wang et al., Nature Reviews, August 2023]
Why it matters: This paper provides a comprehensive look at how the AI revolution is transforming scientific research. As AI capabilities grow more powerful, the ability to accelerate discovery across disciplines ranging from physics to medicine could lead to groundbreaking new insights that benefit human knowledge and wellbeing. However, the authors rightly point out key challenges that must still be overcome to integrate AI safely and effectively into the scientific process. Their analysis helps highlight critical areas for future research and collaboration to ensure AI aids science in broadly beneficial ways.The Zitnik Lab at Harvard has published a remarkable review detailing the role of AI in scientific inquiry and discovery. The piece argues that AI is transforming discovery across sciences from hypothesis generation to data interpretation – it is reshaping all stages of research in ways we could not imagine using traditional methods alone.
The authors highlight recent breakthroughs in AI that are enabling these advances, such as self-supervised learning algorithms that can analyze massive amounts of unlabeled data, and geometric deep learning methods that can capture the structural relationships within scientific data. Powerful generative models can now create new molecular structures, proteins, and other designs by learning from diverse datasets.
According to the authors, AI has already contributed to major feats like solving the 50-year protein folding problem and simulating molecular systems with millions of particles. It is being used across particle physics, materials science, drug discovery, and quantum computing to accelerate research. However, integrating AI into scientific workflows poses new challenges, such as practical barriers like data quality and availability, along with algorithmic limitations in areas like extrapolative prediction and integrating scientific knowledge into models.
Realizing the full potential of "AI for science" will require cross-disciplinary collaboration, advances in theory, methods, and infrastructure, and responsible implementation. If these challenges can be met, AI-driven techniques offer immense opportunities to enhance human understanding through faster, more intelligent, and even fully autonomous scientific discovery. Ultimately, the responsible use of AI can unlock discoveries and insights that were previously unattainable.
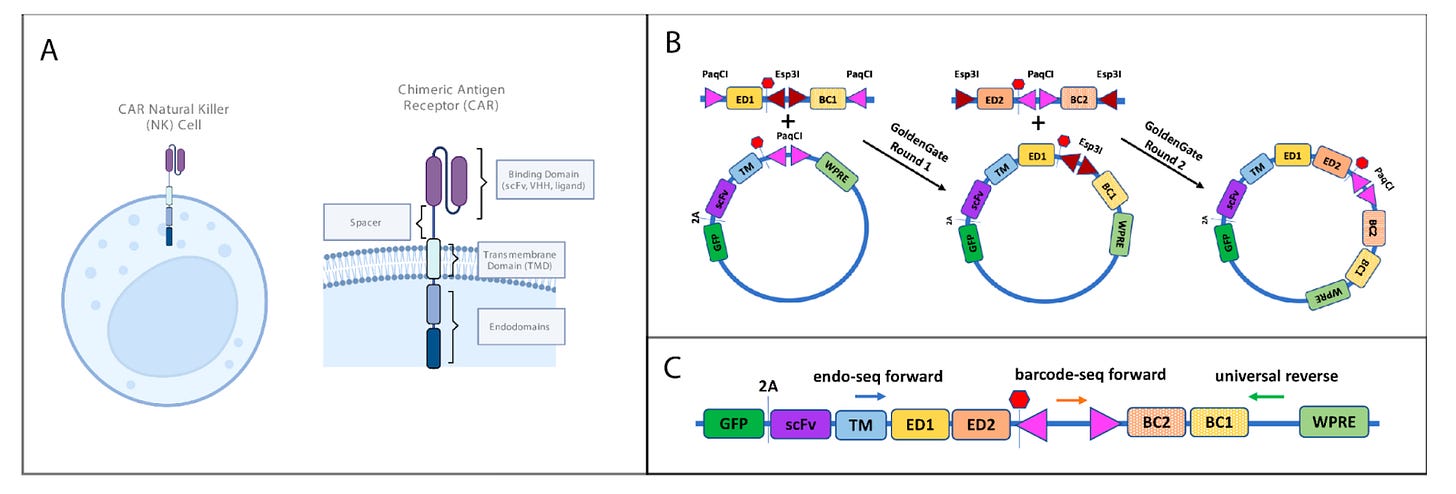
TRAFfic signals: High-throughput CAR discovery in NK cells reveals novel TRAF-binding endodomains that drive enhanced persistence and cytotoxicity [Williams et al., BioRxiv, August 2023]
Why it matters: While the cell therapy field has primarily focused on CAR-T cells, natural killer (NK) possess several advantages. A new high-throughput screening study identifies several new chimeric-antigen receptor (CAR) endodomains that uniquely enhance CAR-NK cell function. Natural killer (NK) cells are a type of cytotoxic lymphocyte that play a crucial role in the innate immune system. Like CAR-T cells, NK cells can be engineered with a chimeric antigen receptor (CAR) to target diseased cells such as cancer. CAR-NK cells possess several advantages over CAR-T cells, including lower risk of graft-versus-host disease and the ability to recognize cancer cells without prior antigen sensitization.
In a new preprint, the team at Modulus Therapeutics, a venture-backed next-gen cell therapy company, developed a high-throughput screening method to identify CAR endodomains (the intracellular part of the CAR receptor that triggers intracellular events leading to cell killing activity; Figure panel A below) that enhanced cell persistence while maintaining cytotoxicity. Libraries were generated of diverse endodomains (Figure panel B) from different signaling pathways. These libraries were then screened for persistence (the ability for cell therapy cells to expand, survive and maintain function) using a serial antigen re-challenge assay, and cytotoxicity was assessed in vitro and in vivo. Several endodomains that significantly enhanced NK cell persistence and cytotoxicity were identified, most notably two TRAF (TNF receptor-associated factor) binding proteins: Fn14 and TANK. Further study is required to determine how these proteins enhance NK cell function, but the big takeaway is that CAR-NK and CAR-T cells are activated by different signaling cascades, and require different endodomain constructs for maximum efficacy.
Immune sensing of food allergens promotes avoidance behaviour [Florsheim et al., Nature, July 2023]
Why it matters: the immune system has wide effects across the body, many of which are unknown. This study examines how the immune system can influence behavior. Specifically interrogating a link between food allergies, the immune system detecting said allergens, and resulting picky eating behavior. Allergies are increasing in prevalence and food allergies in specific may seem like a local interaction between immune cells (mast cells) and the offending allergen stimulus. However, the effects may actually reach much farther. This study suggests that there is communication between the brain and the immune system which leads to avoidance of the offending food when food allergies are concerned.
The mechanism of action, though poorly understood, is thought to involve food allergens interacting with mast cells in the epithelial lining of the gut. These cells bind the IgE antibodies associated with recognizing the allergen. Interestingly, re-exposure to the allergen causes mast cells to release leukotrienes and the epithelial cells to release the GDF15 protein. This signals to a few regions of the brain (nucleus of tractus solitarius, external lateral parabrachial nucleus, and the amygdala) to change behavior. Based on this mechanism, it is not out of the question that you can develop an allergic response to a food without the actual stimulus being present, so long as the requisite factors are released for behavior to be altered. Mice, when given the choice between water with allergen and water without, showed avoidant behavior towards the allergen-containing water. If interested in this topic, another study with parallel findings was also published last month.
What we listened to
RAAIS 2023 - Multimodal Medical AI [Vivek Natarajan, Research Scientist at Google Health AI]
Notable Deals
Steve Jobs’ son raises $200M for cancer-focused venture fund
In case you missed it
AI for Brain-Computer interface [Github]: A curated list of resources dedicated to AI (ML/DL/RL) applied to brain-computer interfaces.
Accelerating Drug Discovery and Development: Synergy Between Wet and Dry Lab [Nima Ronaghi, Breakout Ventures, August 2023]
Field Trip
Did we miss anything? Would you like to contribute to Decoding Bio by writing a guest post? Drop us a note here or chat with us on Twitter: @ameekapadia @ketanyerneni @morgancheatham @pablolubroth @patricksmalone