BioByte 042: 36 billion compounds, genes that keep you alive, spatially-resolved sc-translatomics, predicting the effect of genetic variants on human proteins, myelin dynamics in multiple sclerosis
Welcome to Decoding Bio, a writing collective focused on the latest scientific advancements, news, and people building at the intersection of tech x bio. If you’d like to connect or collaborate, please shoot us a note here or chat with us on Twitter: @ameekapadia @ketanyerneni @morgancheatham @pablolubroth @patricksmalone. Happy decoding!
About to get kicked off a plane? Seconds away from being arrested? Hope you lock this down cause you never know what fun facts may impress the judge:
A deep dive into screening 36 billion compounds with Recursion
A look into the “unknome” - genes that may contribute to keeping you alive
A lesson from Rich Sutton on how computation will eventually surpass human capabilities
RIBOMap - a new method for spatial mapping of protein synthesis at single-cell and subcellular resolution
A new deep protein language model, ESM1b, which can predict the effects of genetic variants on human proteins
A new paradigm in multiple sclerosis - how myelin encapsulating axons may actually increase the risk of degeneration in disease
Bringing the kidney back in vogue: using AAV for gene therapy of nephrotic syndromes
What we read
Blogs
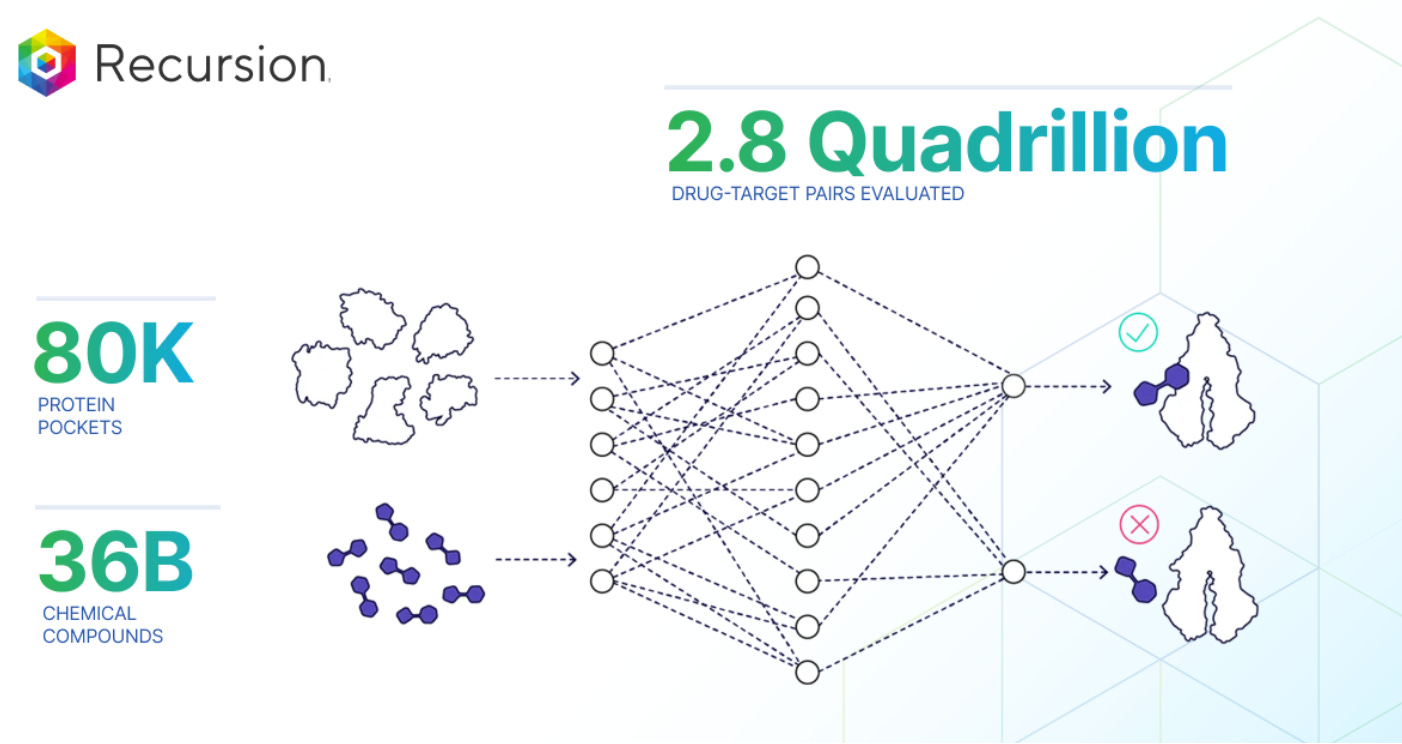
A Deep Dive into Screening 36 Billion Compounds: Q&A with Stephen MacKinnon [Recursion, 2023]
Recursion carried out an ultra-large virtual screen using the Enamine REAL Space library (36B chemical compounds) against the structure of 80K individual protein pockets (both experimentally-determined and modeled). Cyclica’s GPU-adapted MatchMaker model, recently integrated into Recursion after the company’s acquisition, made the simulation possible.
The team claims that this drug-target interaction (DTI) prediction model will have the following advantages: “first, this predicted data layer can be used to determine which wet-lab experiments should be executed to advance programs faster across a wide range of targets and chemical space. Second, this predicted data layer can be used as part of Recursion’s multi-modal dataset to better understand biological activity across programs quickly and at scale. Finally, this approach can pre-screen for more computationally expensive precision modeling techniques implemented by Recursion’s computational and digital chemistry teams, to more efficiently advance programs.”
Whilst the technology and the scale of the prediction is interesting to see, we’d love to see a quantitative measure of its utility. These metrics could include model accuracy of predicted interaction pairs, decrease in cost given that Recursion should be able to choose experimental starting points better or the marginal benefit of this new data set when combined with its phenomics database.
The Mystery Genes That Are Keeping You Alive [Highfield, Wired, August 2023]
Despite significant advances in functional genomics (a field of molecular biology that aims to describe gene functions and interactions), the function of the much of the genome (the “unknome”) remains unknown. The human genome encodes 20k proteins, and a new study estimates that the function of ⅕ of these genes is still not elucidated. To arrive at this estimate, the authors assessed how conserved a particular gene is across species (genes that remain unchanged in many species are likely to have important functions), and then scored each gene according to how much was known in the literature about its function. Low-scoring genes corresponding to genes with unknown functions were then knocked down in a Drosophila model, and surprisingly often had lethal effects on the organism, suggesting that these unknown genes had fundamental functions for survival. The authors published the “Unknome” in a publicly available database.
The Bitter Lesson [Rich Sutton, March 2019]
We were recently sent this essay by Rich Sutton, the father of reinforcement learning, from four years ago on AI research that is a powerful enough read to warrant the reminder. The first line of the essay captures the bitter lesson itself–”general methods that leverage computation are ultimately the most effective, and by a large margin”. Sutton comments on the tradeoff between computation-led performance improvements and those based on human-knowledge. In the short term, human knowledge might prove powerful. But over time and when you take Moore’s law into consideration, computation will win; it is less messy and more generalizable. He provides several examples of where we made this mistake (computer chess, computer Go, speech recognition, computer vision etc.) and boils the seventy years of lessons into the following takeaway:
1. AI researchers have tried to build knowledge into their agents
2. This always helps in the short term and is personally satisfying to the researcher
3. In the long run it plateaus and even inhibits further progress
4. Breakthrough progress eventually arrives by an opposing approach based on scaling computation by search and learning There’s a secondary lesson here that is particularly thought provoking for biology. If “the actual contents of minds are tremendously, irredeemably complex” the same can be said of biological systems (of which we barely understand even a little bit of). How do we capture this complexity and approximate systems to understand and discover biological insights?
Sci-Fi Idea Bank [Packy McCormick, Not Boring, August 2023]
Packy is back at it with another optimistic post of ideas. He put together a list of >3,000 sci fi ideas, operating under the thesis that ideas for new technology appear in science fiction before they become science reality. For example, he cites that Gulliver’s Travels, published in 1726, references 3D modeling, search engines, biofuels, and floating rocks…In fact, he makes the argument that it is hard to find a counterexample where a now powerful technology did not first appear in sci-fi. And because biotech has historically been portrayed in a negative light in science-fiction and media, this list is a chance to see all the world-positive things you can do with science and biology. There are far too many ideas to parse through but some highlights as it pertains to the life sciences:
Express dolphin—locomotion under the waves
Compact food pastilles—one small tablet to be a month’s worth of food
Rainmaker—specific way to produce rain on demand
Diaper that indicates when it is wet
Pill that counteracts the effects of fatigue
Chemical artificial intelligence
Light bulb that could sustain life without sun
Academic papers
Spatially resolved single-cell translatomics at molecular resolution [Zeng et al., Science]
Why it matters: To fully understand translational gene regulation, it is crucial to measure protein synthesis, yet studying mRNA translation at scale with spatial and single-cell resolution remains a challenge. Zeng et al., introduce RIBOmap, a multiplexed method for spatial mapping of protein synthesis at single-cell and subcellular resolution.The methodology named ribosome-bound mRNA mapping (RIBOmap) uses a targeted-sequencing strategy in which tri-probes selectively detect and amplify ribosome-bound mRNAs. The tri-probe set includes a:
Splint DNA probe: hybridizes to rRNAs and circularizes padlock probe
Padlock probe: targets specific mRNA species of interest and encodes a unique barcode
Primer probe: targets mRNA site adjacent to the one targeted by the padlock probe and serves as the primer for RCA resulting in an amplicon
Previous research has focused on using mRNA levels as a proxy for protein abundance, which has been revealed to have a poor correlation given subcellular-localized translation. RIBOmap can be added as an effective tool to understand translation independent of circulating mRNA and protein abundances.
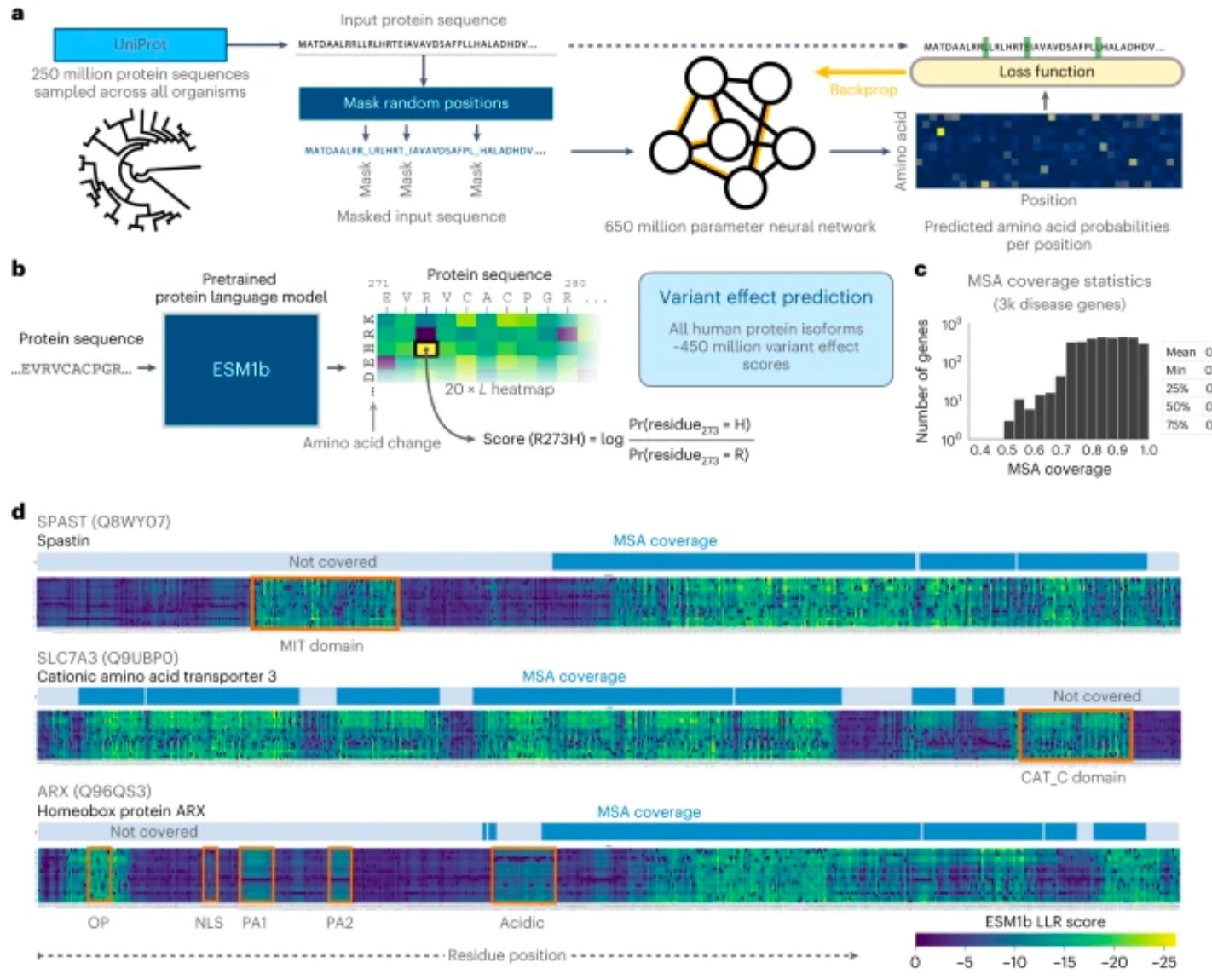
Genome-wide prediction of disease variant effects with a deep protein language model [Brandes et al., Nature Genetics]
Why it matters: A new deep protein language model called ESM1b demonstrated striking performance in predicting the effects of genetic variants on human proteins. ESM1b was trained on a massive dataset of protein sequences and their corresponding functions, and it can predict the effects of both single-nucleotide variants (SNVs) and small insertions or deletions (indels).The researchers evaluated ESM1b on a variety of benchmark datasets, and it consistently outperformed other state-of-the-art methods for predicting variant effects. For example, on the ClinVar benchmark, which contains a set of variants with known clinical significance, ESM1b achieved an accuracy of 88%, compared to 78% for the previous best method.
The development of ESM1b is a significant advancement in the field of genomics. It provides a powerful new tool for predicting the effects of genetic variants and could be used to identify individuals who are at risk for developing genetic diseases, develop new diagnostic tests for genetic diseases, and design new drugs and therapies.
Myelin insulation as a risk factor for axonal degeneration in autoimmune demyelinating disease [Schäffner et al., Nature Neuro]
Why it matters: Multiple sclerosis (MS) is classically thought of as a demyelinating disease, where the amount of demyelination correlates with disease severity and progression. A new study suggests that myelin encapsulating axons actually increase the risk of axon degeneration in MS, with important implications for next-gen remyelination therapies. MS is an autoimmune neurological disorder in which immune cells attack oligodendrocytes, glial cells in the central nervous system that insulate neuronal axons with myelin to improve signal conduction between neurons. A common therapeutic strategy is to prevent further demyelination of axons to arrest disease progression. A recent study in Nature Neuro complicates this strategy by showing that myelin ensheathment can itself actually become detrimental to axonal survival in an inflammatory environment. The authors showed that axons with permanent damage almost always contained myelin, while axons without myelin were spared from degeneration. The likely explanation is that once oligodendrocytes are damaged, they lose metabolic support and shift from a protective and potentially harmful state for axons. These findings have important implications for the next-generation of MS therapies that promote remyelination. Rather than stabilizing damaged axons and myelin, a better therapeutic strategy will be to promote remyelination and restoration of normal oligodendroglial function.
Adeno-associated virus gene therapy prevents progression of kidney disease in genetic models of nephrotic syndrome [Ding et al., Science Translational Medicine]
Why it matters: Gene therapy for kidney diseases has remained stagnant, with the largest bottleneck being in the specific delivery of genetic cargo. In this paper, the authors develop an AAV-based gene therapy to treat two different mouse models of nephrotic syndrome, bringing tailwinds for novel treatments of kidney disease. Despite the significant advances in gene therapy to date, there has been no success story in treating monogenic kidney disease. Although studies over the years have demonstrated success in AAV-mediated transduction of the kidney, these largely affected the tubular epithelium (lining the nephrons), with minimal success in modifying other cells including the podocytes (barrier-like filtration cells), which are heavily implicated in the nephrotic (protein-wasting) syndromes. Given some of these nephrotic syndromes (such as childhood steroid-resistant nephrotic syndrome) are genetically driven (with mutations in genes such as NPHS2, which encode for podocin, a critical part of the kidney’s filtration capabilities), these represent potentially tractable diseases for gene therapy.
The authors identified AAV-LK03 (closely related to AAV3) as a serotype that highly transduced human podocytes and proximal tubular cells in human cells. They showed that delivering wild-type podocin rescued podocyte adhesion derangements. However, AAV-LK03 is known to poorly transduce mouse cells, and so they used AAV2/9 for in vivo mouse work. To ensure podocyte-directed expression, the authors evaluated a few promoters (including CMV and nephrin promoters) and found that the minimal human nephrin promoter led to improved efficacy. Using these, the authors were able to demonstrate that gene transfer in both an inducible podocin knockout and knock-in model successfully rescued kidney disease. Although work still remains on identifying optimal promoters to minimize off-target expression (which was seen in the liver and spleen), this work brings gene therapy for kidney disease back into the foray.
What we listened to
Notable Deals
Novo adds another obesity drug in $1B deal for startup Inversago
Alltrna raises $109M for ‘transfer RNA’ drug vision
Maryland biotech Georgiamune nabs $75M to fund three cancer, autoimmune clinical trials
RNA startup ADARx raises $200M from Bain, TCGX to bring RNAi 'to the next level'
Merck enlists Astex to search for p53 cancer drug in expanded deal
Agios to take over Alnylam’s preclinical siRNA blood disorder asset in a deal worth $147.5M
In case you missed it
What we liked on Twitter
Field Trip
Did we miss anything? Would you like to contribute to Decoding Bio by writing a guest post? Drop us a note here or chat with us on Twitter: @ameekapadia @ketanyerneni @morgancheatham @pablolubroth @patricksmalone