BioByte 043: new implications for schizophrenia gene, what characterizes a cell type, multi-kilobase RNA editing, why age of menarche has dropped six years
Welcome to Decoding Bio, a writing collective focused on the latest scientific advancements, news, and people building at the intersection of tech x bio. If you’d like to connect or collaborate, please shoot us a note here or chat with us on Twitter: @ameekapadia @ketanyerneni @morgancheatham @pablolubroth @patricksmalone. Happy decoding!
Brain fried from that end of summer heat? We’ve got you covered with the highlights:
Schizophrenia gene found to be linked to energy dysfunction in brain cells
What characterizes a cell type and more on building a cell atlas
Breaking down preclinical drug discovery
Large-scale multi-kilobase RNA editing
GLP-1 inhibitors and their influence on neural reward pathways in obesity
Autoimmune cause of CNS-restricted vitamin B12 deficiency
Barcoded pooled CRISPR screening for phenotypic drug discovery
Why the average age of menarche has dropped six years
What we read
Blogs
Schizophrenia gene found to be linked to energy dysfunction in brain cells [Isabella Cueto, STAT, 2023]
Schizophrenia has been known to be heritable for over a century, yet the biological mechanism of action that links genotype to the clinical phenotype is poorly understood. In the early 2000s, Jennifer Mulle found that the 3q29 deletion is associated with a 40-fold increase in the risk of developing schizophrenia, making it the strongest individual genetic risk factor for the disease. Its rarity, however, means the deletion only explains 1 of 1,000 cases of schizophrenia.
In her latest work, Mulle offered an explanation to how this deletion leads to schizophrenia. They noticed that the mitochondria in cells with the deletion lacked ‘metabolic flexibility’, which meant they could not shift between respiration modes leading to critical effects in neuron differentiation and maturation. Whilst the exact genes within the deletion that drive this effect need to be identified, this extensive study could indicate that broader neurodevelopmental disorders arise due to genetic aberrations that affect cell metabolism.
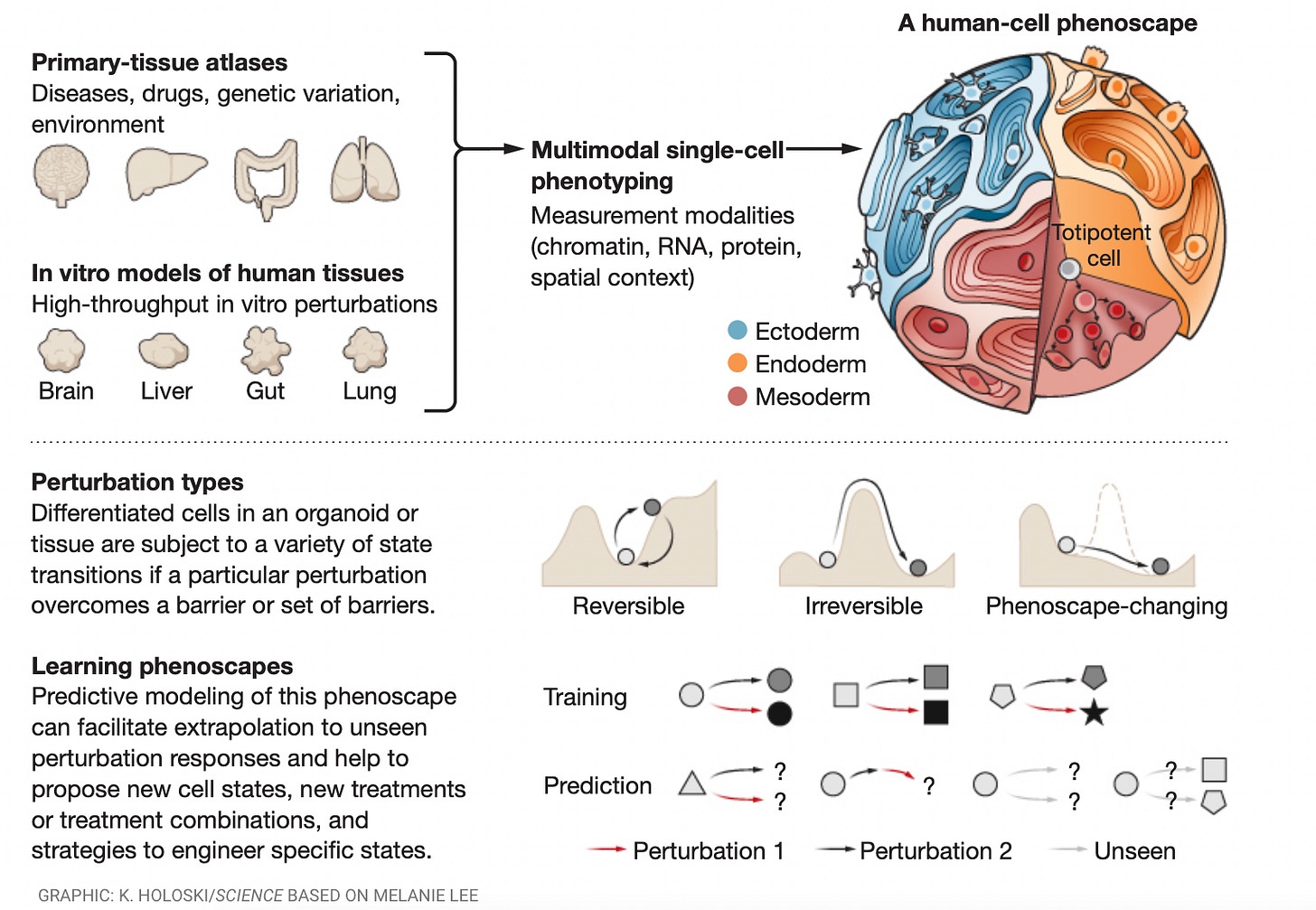
What is a cell type? [Fleck et al., Science, August 2023]
There are a number of research efforts focused on generating atlases of cell types on the basis of molecular features like the transcriptome, epigenome, and proteome, as well as the various cell states for a given cell type. An emerging view is that an important component of a cell’s state is how it responds to a genetic, chemical, or environmental perturbation. Cells can vary widely in response to perturbations such as genetic mutation or pathogen infection. The landscape of all possible cell states in response to perturbations is called a cell’s phenoscape. A key bottleneck in the generation of a phenoscape map, however, is a paucity of biological data that causally demonstrates cellular responses to environmental changes. To address this limitation, perturbations need to be induced and cell responses characterized under controlled experimental conditions in high throughput in in vitro model systems, ideally in 3D culture systems such as organoids that more accurately recapitulate the in vivo tissue microenvironment.
The Backbone of Drug Discovery [Vega Shah & Sanjay Saraf, Bits in Bio, August 2023]
Vega and Sanjay took to the Bits in Bio substack to put out a comprehensive overview on drug discovery, walking through target identification, what goes into picking a good target, target validation, computational techniques, and how things are done today. This is a must read for anyone new to the drug discovery world, anyone who needs a refresher, or anyone who is just curious about what happens during the years of preclinical development that go into making a therapeutic. The examples are excellent! Let us know if you find a similar overview for what happens afterwards in clinical development.
Does Image Reconstruction Matter in Drug Discovery [Chris Gibson, August 2023]
Chris Gibson recently shared more on Recursion’s foundation models that are being built for image reconstruction—in other words, can a model be trained on biological image data to predict missing parts of an image (read: predict biological phenotypes). He took to this quick post to clarify what Recursion’s models are doing and why this type of information is relevant. The tl;dr here is such models can make sense of cell perturbations from a biological and chemical standpoint making it easier to figure out what perturbations may be needed to restore diseased cells or achieve a given state based on where the cell is today. Almost like a cellular map. Now it’s important to remember many groups are working on this with different approaches. Recursion’s image-based approach is one way to attempt to crack the problem of creating a biological map.
Academic papers
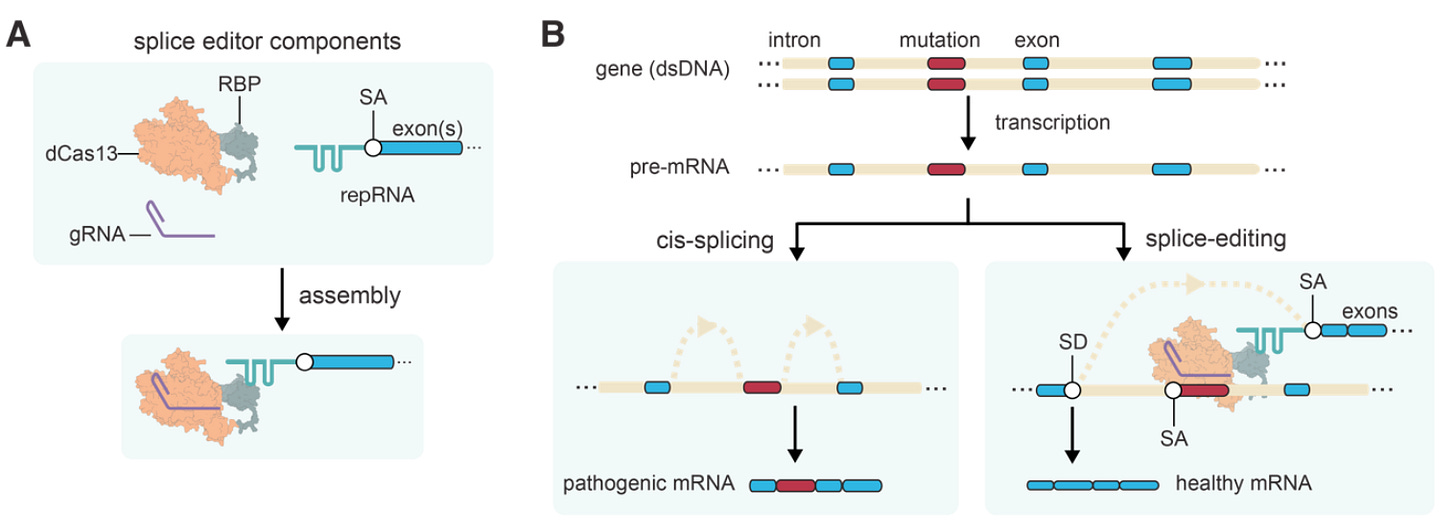
Programmable multi-kilobase RNA editing using CRISPR-mediated trans-splicing [Borrajo et al., bioRxiv, 2023]
Why it matters: Most gene editing approaches in the clinic or in preclinical development are limited to single base edits (e.g. ADARs, dCas base editors) or small DNA insertions and deletions (e.g. CRISPR/Cas9, ZFNs) and their adoption is limited due to unintended off-target effects or challenges in the delivery of large cargo. Large-scale RNA editing can overcome these challenges and expand the treatable disease population.The team at Amber Bio have developed a CRISPR-mediated Splice Editor, which enables large-scale insertions in RNA by co-opting the endogenous RNA-splicing machinery (spliceosome). The results demonstrate an efficient, precise and programmable editing method for protein correction which can be used to expand the treatable population for monogenic diseases with large allelic diversity without permanent off-target effects of DNA editing.
Following transcription, eukaryotic cells transform pre-mRNA into mature mRNA, which is the final molecule used by ribosomes to produce proteins. Part of that process includes ligating exons and excising introns from the pre-mRNA, which is carried out by the spliceosome, by joining a splice acceptor with a splice donor. The Splice Editor, pictured above, is designed to carry out trans-splicing; this is when a single chimeric mature mRNA molecule is formed from more than one pre-mRNA transcript. The editor is composed of gRNA, a dCas13 fused to an RBP and a repRNA (which contains an exon of interest).
The advantages as a therapeutic modality include i) that it doesn’t rely on endogenous DNA repair pathways which can limit editing in post-mitotic cells due to its inefficiency, ii) can offer a gene editing solution for diseases whose gene size exceeds the packaging limit of AAV, iii) dose is self-regulated by endogenous translation rate, which increases safety and iv) can correct numerous mutations using one therapy, rather than target single mutations at a time, leading to some rare allele carriers left behind.
Liraglutide restores impaired associative learning in individuals with obesity [Hanssen et al., Nature Metabolism, August 2023]
Yet more evidence of the diverse and surprising physiological effects of GLP-1 inhibitors in the treatment of obesity. In this study, patients on a GLP-1 inhibitor underwent functional MRI imaging while performing an adaptive learning task. Sensory learning was impaired in patients with reduced insulin sensitivity, and learning was restored with GLP-1 inhibitor treatment. Analysis of the fMRI data suggested that dopaminergic midbrain regions as the site of action. These findings suggest a potential mechanism by which GLP-1 inhibitors could alter neural processing to influence food-seeking behavior and food-related reward in obesity.
Transcobalamin Receptor Autoantibodies in Central Vitamin B12 Deficiency [Pluvinage et al., medRxiv, 2023]
Why it matters: Vitamin B12 (cobalamin) is a critical vitamin critical for DNA synthesis, playing key roles in hematopoiesis and myelination. Vitamin B12 deficiency can lead to a number of conditions, including anemia, intestinal malabsorption, and a suite of neurological issues including reversible dementia and subacute combined degeneration of the spinal cord. In this paper, the authors discover an autoimmune cause of CNS-restricted vitamin B12 deficiency, due to an autoantibody targeting the transcobalamin receptor (CD32). This work identifies a new form of metabolic neurologic disease that may be responsive to immunomodulation and vitamin supplementation.A 60 year old female presented to the hospital with a number of neurological complaints, including bilateral lower extremity pain, dysarthria, and tremors. A series of work-up demonstrated lesions in her cerebellum, while laboratory measurements were largely unremarkable. She was treated with steroids, which led to an improvement in her neurological exam over 3 months; eighteen months later, she was diagnosed with lupus and treated with the antirheumatic drug, hydroxychloroquine. Over time, her initial neurologic deficits stabilized, while she experienced worsening word-finding difficulty. Of note, her MRI showed she still had persistent cerebellar lesions, and her serum vitamin B12 levels were normal.
Using phage immunoprecipitation sequencing (PhIP-seq), the authors were able to screen for autoantibodies against a library of >700K peptide sequences, and found an autoantibody targeting the B12 transporter (CD320). The patient above had nearly undetectable levels of B12 in her CSF. Beyond her, the authors found anti-CD320 autoantibodies in 7 other cases (of neurologic deficits of unknown etiology), all of which targeted the same epitope. Subsequently, they screened 85 grad students and found anti-CD320 autoantibodies in 6% of that population; additionally, they analyzed a cohort of 132 samples from multiple sclerosis patients and similarly found a 6% seropositivity rate. This work is exciting as it reinforces how autoimmunity remains such an unplumbed domain, and plays a critical role across disease manifestations. Plenty of questions still remain about anti-CD320 autoantibodies and their role in CNS B12 deficiency; however, these findings elucidate a previously unknown cause of neurologic disease.
Every so often, we’ll feature a guest submission from someone in the community to expand the papers we cover. If you read a really cool paper and want the chance to be included in next week’s newsletter, email us with your commentary and writeup following the format above at decodingbio@gmail.com. This week we hear from Sidharth Sirdeshmukh:
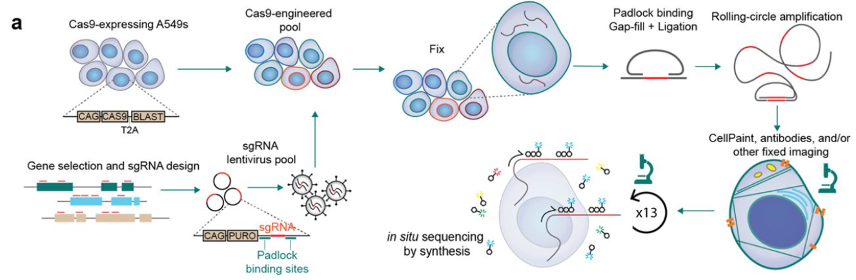
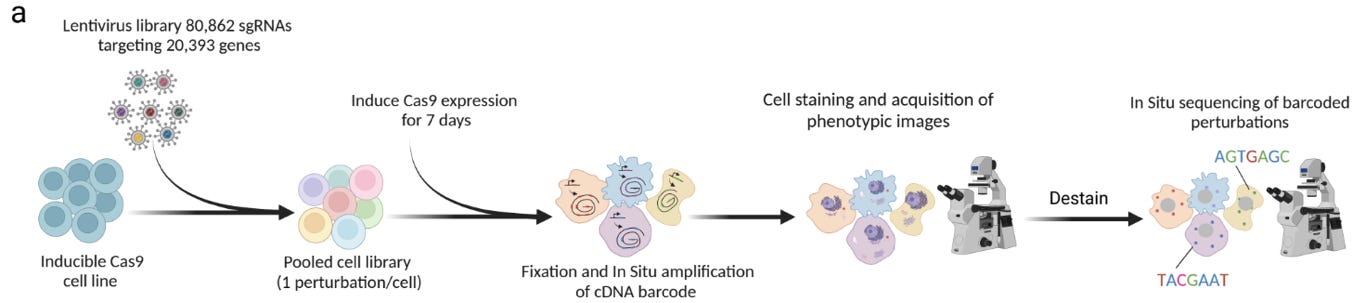
High Throughput Pooled Optical Screening: POSH and PERISCOPE
Why it matters: Traditional CRISPR screening is a powerful method that allows researchers to infer pathway-level interactions from single gene knockouts. Teams at Insitro and the Broad Institute (Carpenter-Singh Lab) have developed highly similar approaches that combine barcoded pooled CRISPR methods with object-level cell imaging to link genetic perturbations to cell morphology. These approaches could be adapted/ improved by any AI/ML-enabled phenotypic drug discovery company to develop internal reference databases for MOA prediction.Here is a snapshot of some of the similarities and differences between both papers:
Wet lab: Both teams used 6-well plates and microscopes from the same manufacturers. They also ran cell fixation, reverse transcription of sgRNAs, Cell Painting (staining and imaging), and in-situ-sequencing via imaging in the same order. The biggest differences were in the design of cell lines, imaging probes, and the focuses of their sgRNA libraries. The intro figures below tell most of the story (Insitro (POSH) on top, Broad (PERISCOPE) on the bottom):
Dry lab: Insitro specifically used deep learning methods for cell segmentation, barcode identification, and cell morphology feature extraction, and have some figures on the predictive accuracy of their models. The Broad on the other hand ran traditional Cell Profiler to extract conventional annotated features like intensity, granularity, and texture to calculate morphological signal scores, which they used to run GSEA analysis with the barcoded knockouts.
For an in-depth comparison of the methods and commentary on the relevance of this research for phenotypic drug discovery, please see the in depth post on Back To The Lab.
What we listened to
Notable Deals
Genesis Tx raises a $200M Series B
Moderna and CARsgen partner to advance a CAR-T cell therapy and cancer vaccine combination
Abcura raises $155M to take rare muscle disease antibody tx through phase 2 and 3
Talus announces $4.3M in grant funding to advance pediatric cancer therapeutics
In case you missed it
The unrealized promise of embryonic stem cells some 25 years later , MIT Technology Review
What we liked on Twitter
Field Trip
not the music you came for but hey, it’s Costco
Did we miss anything? Would you like to contribute to Decoding Bio by writing a guest post? Drop us a note here or chat with us on Twitter: @ameekapadia @ketanyerneni @morgancheatham @pablolubroth @patricksmalone










Thanks for flagging Jennifer Mulle's work on decoding the connections between genetics and Schizophrenia! I need to dig into more of her previous work ⛏
In the article about the new form of B12 deficiency, the authors remind the reader of cerebral folate deficiency, a similar condition. Lisa Pan et al. recently (2022) reported on a replication study of patients with severe treatment-resistant depression, in which some 5% of them were found to have cerebral folate deficiency.