BioByte 046: pan-cancer AI models, benchmarks for single-cell LLMs, emergent properties of chemical language models, new astrocyte subtype uncovered
Welcome to Decoding Bio, a writing collective focused on the latest scientific advancements, news, and people building at the intersection of tech x bio. If you’d like to connect or collaborate, please shoot us a note here or chat with us on Twitter: @ameekapadia @ketanyerneni @morgancheatham @pablolubroth @patricksmalone. Happy decoding!
Late but great issue this week. Easing into fall, curl up and join us for a recap on all things bio x AI this week:
Pan-cancer AI histopathology model under development by Microsoft and Paige AI
Genetic fine-mapping and multi-omics data bring fresh perspectives to understanding hypertension
Benchmarks and design guidelines for applying LLMs to single-cell data
Chemical language models unlock exploration of both chemical and protein space simultaneously
Novel astrocyte subtype involved in memory and movement neuronal circuitry uncovered
What we read
Blogs
Microsoft and Paige are building the world’s largest AI model for detecting cancer [CNBC, September 2023]
Microsoft has partnered with digital pathology provider Paige to create the world's largest image-based AI model for detecting cancer. This AI model, trained on an unprecedented amount of data, can identify both common and rare cancers, potentially aiding doctors dealing with staffing shortages and increasing caseloads. Paige has been working to digitize pathology workflows, and their FDA-approved viewing tool, FullFocus, allows pathologists to examine digital slides on screens instead of microscopes. However, digital pathology comes with significant storage costs, which Paige aims to overcome with Microsoft's help.
Paige's AI model is currently the largest publicly announced computer vision model, trained on 4 million slides to identify various cancer types. The partnership between Paige and Microsoft aims to enhance cancer care without replacing pathologists, and their research will be published through Cornell University's preprint server arXiv, with future steps including thorough testing and regulatory collaboration before product rollout.
The next-generation CAR-T therapy landscape [Verma et al., Nature Drug Discovery Reviews, 2023]
CAR-Ts have unequivocally become an important treatment for cancer due to their outstanding efficacy for blood cancers. Since 2017, six therapies have been approved and there are >200 assets currently in clinical trials.
The modality, however, has been limited by safety (e.g. high rates of adverse events, CRS), efficacy (e.g. inability to overcome TME, cell exhaustion) and operational hurdles (e.g. capacity constraints, extensive patient monitoring, lengthy manufacturing).
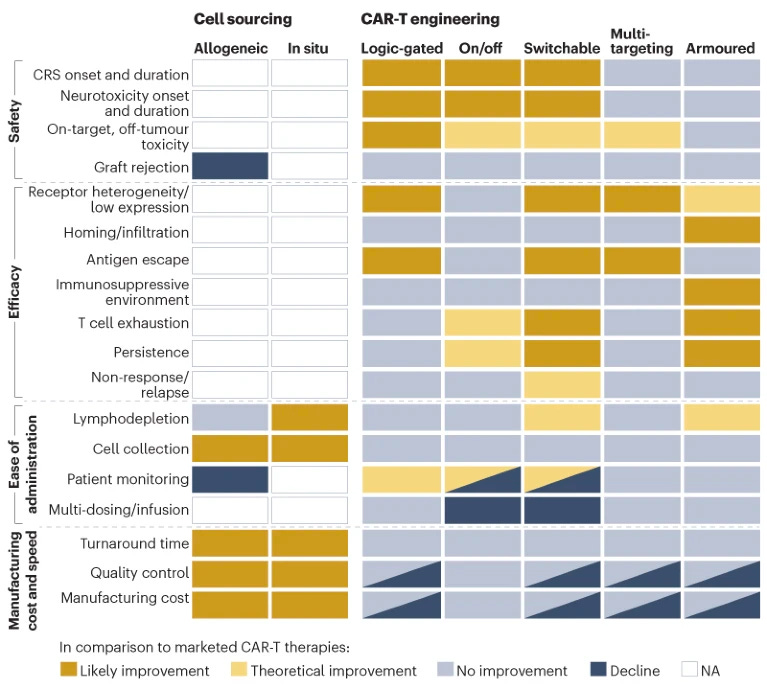
In order to address these limitations, the new generation of CAR-T therapies use different cell sources and engineering approaches. The authors of the review created the heatmap below which shows the effects of each modification on safety, efficacy, administration and manufacturing.
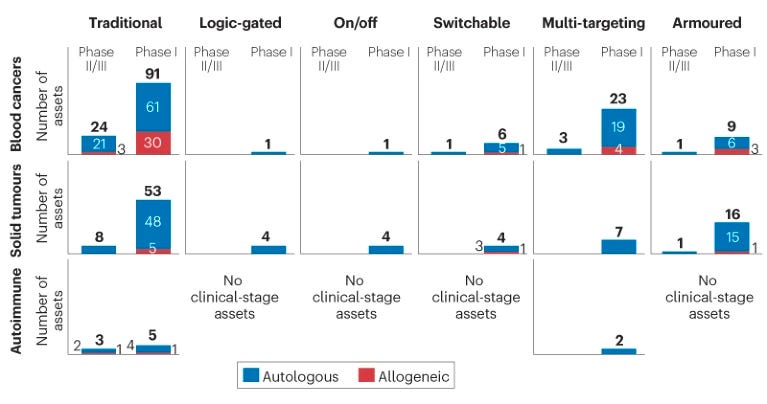
In the clinic, most assets are still using the single-antigen autologous approach (as seen in the diagram below). However, there are many CAR-T assets using armoured, switchable and multi-targeted approaches advancing into the clinic.
Sterile Information: Early Forecasting Not the Answer to R&D Productivity Woes [David Shaywitz, Timmerman Report, Sep 2023]
David Shaywitz discusses in the Timmerman Report decision-making in biopharma organizations, and balancing commercial considerations and market opportunities with the quality of science when prioritizing drug programs, and. Novartis recently has restructured to tip the balance in favor of commercial vs scientific factors in early-stage R&D. Yet, predicting peak sales of a drug is tricky, and studies have shown little correlation between the predicted sales at time of approval vs actual peak sales of a drug. An alternative is to prioritize programs with good translational models that give the drug developer confidence that positive preclinical data will translate to clinical success.
A secondary question is whether novel drugs will emerge from startups or large pharma. Pharmaceutical giants like Merck and Millenium have seen unexpected successes with drugs initially overlooked. While big pharma possesses vast resources, its bureaucratic and process-driven nature can stifle innovation. True drug discovery requires agility, imagination, and the ability to embrace uncertainty—qualities often absent in large corporations but present in nimble startups. As a result, the future of groundbreaking drug discoveries might lean more toward startups than traditional big pharma.
Academic papers
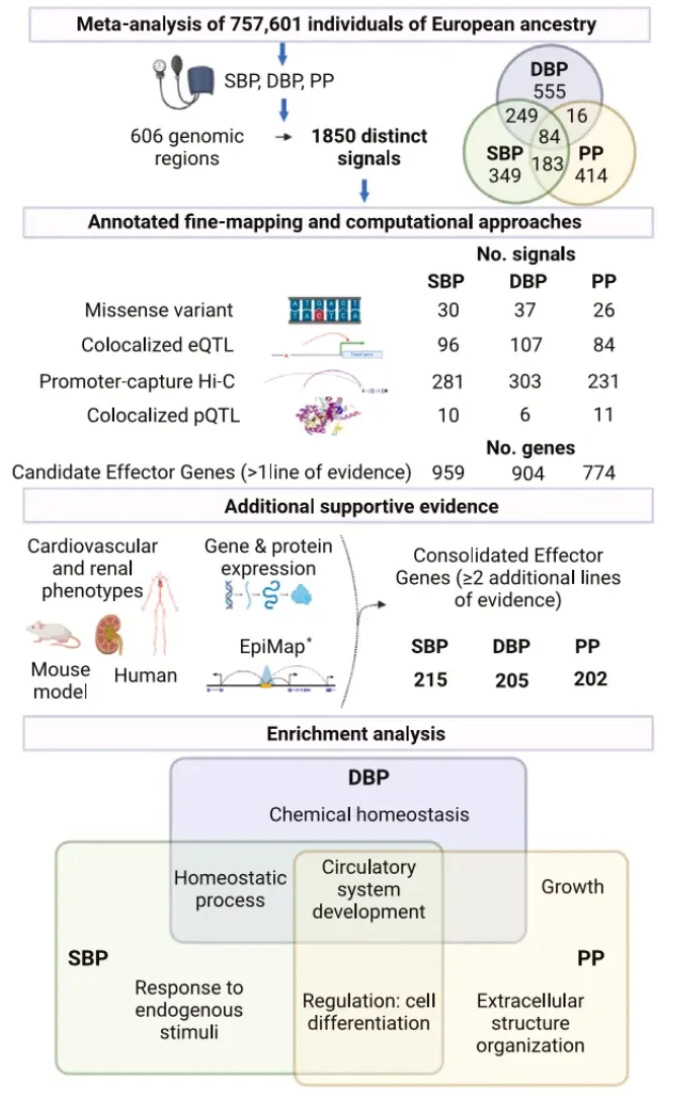
Integration of genetic fine-mapping and multi-omics data reveals candidate effector genes for hypertension [Stefan van Duijvenboden et al., Cell, September 2023]
Why it matters: High blood pressure is a major risk factor for cardiovascular disease worldwide, but the genes and pathways that influence blood pressure are still not fully defined. By consolidating evidence from large genetics datasets and tissue-specific molecular profiling, these researchers have generated a priority list of hundreds of causal genes for experimental validation. Characterizing these genes and the mechanisms through which they act will provide insights into blood pressure regulation. This could ultimately identify novel drug targets or repurposing opportunities for improved treatment of hypertension. The multi-tiered computational approach also serves as a model for dissecting the genetics of other complex diseases.This study integrated multiple genomic and molecular profiling datasets to identify potential causal genes influencing blood pressure regulation. The researchers started with published genome-wide association data on three blood pressure traits in over 750,000 people. Through statistical techniques and comparison with tissue-specific functional genomic annotations, they localized distinct association signals and prioritized likely causal variants. These included coding variants in established (like SLC39A8 and ADRB2) and novel genes.
By integrating data on gene expression and chromatin interactions in disease-relevant tissues, they predicted target genes of regulatory variants. Combining evidence from multiple analyses highlighted 436 high-confidence candidate "effector" genes, including known players like AGT and ACE as well as newly implicated genes. These genes were enriched in pathways like circulatory system development and kidney function.
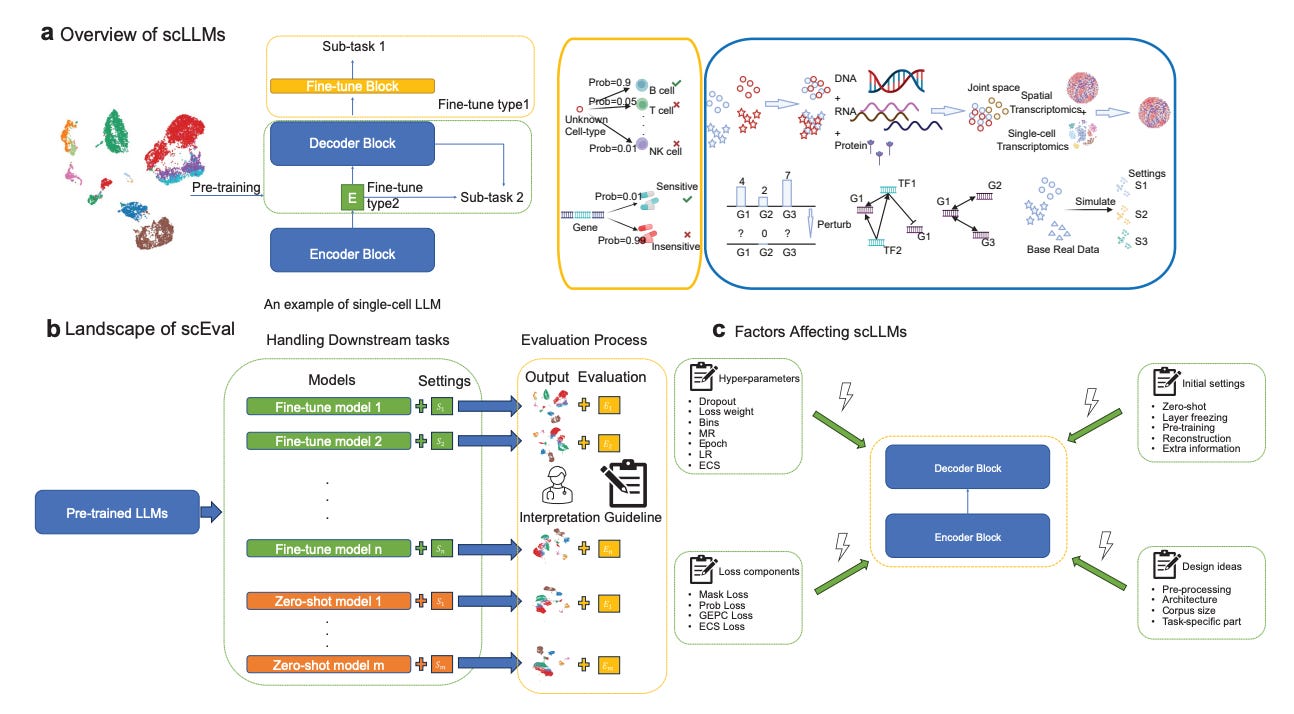
Evaluating the Utilities of Large Language Models in Single-cell Data Analysis [Liu et al., bioRxiv, September 2023]
Why it matters: This analysis provides valuable benchmarks and design guidelines as researchers increasingly look to adapt the successful LLMs from natural language processing to single-cell biology. Characterizing the capabilities and limitations of current single-cell LLMs will inform efforts to develop more versatile foundation models for this domain. With optimized model training, single-cell LLMs could become powerful tools for extracting insights from the wealth of single-cell data. More robust LLMs could enhance interpretation of cell atlases and elucidation of disease mechanisms to advance medicine. This study offers an important step toward realizing the potential of LLMs in single-cell research.In this study, researchers evaluated the performance of several large language models (LLMs) designed for single-cell data analysis across eight key tasks. The LLMs included scGPT, scBERT, Geneformer, and others. Through experiments on datasets spanning cell type annotation, batch effect correction, perturbation prediction, and more, the authors found LLMs can excel at certain tasks like cell type classification but lag behind specialized methods in other areas. Hyperparameter tuning and task-specific loss functions were critical for optimizing LLM performance. LLMs showed promising emergent abilities like cross-species transfer learning for cell typing. However, stability across datasets and tasks needs improvement.
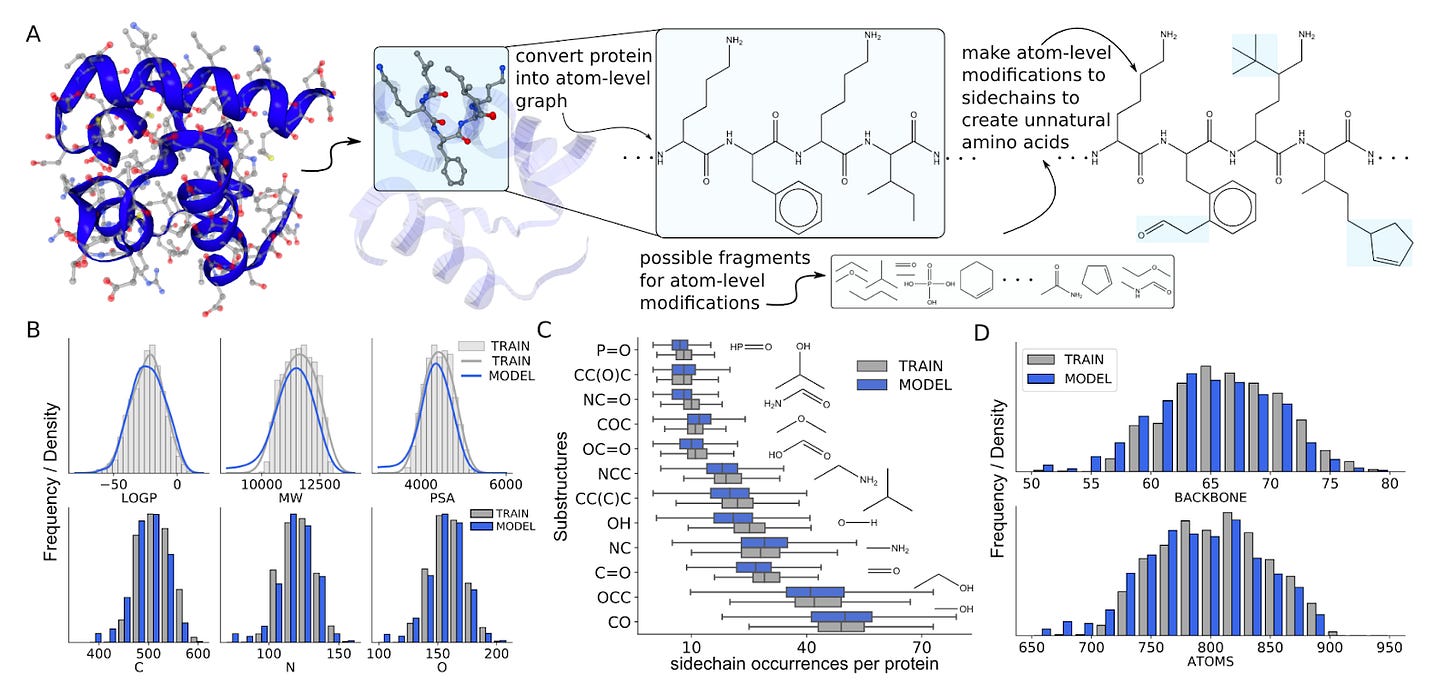
Atom-by-atom protein generation and beyond with language models [Flam-Shepherd et al., aRxiv, 2023]
Why it matters: Large language models used to generate proteins have largely been used at the amino acid scale (biological models), rather than at the atomic scale (chemical models). As seen in this paper, using chemical language models for protein generation enable the generation of proteins with amino acid modifications (unnatural amino acids) and the exploration of both chemical and protein space at the same time (for example, through the generation of antibody-drug conjugates).This paper from the University of Toronto and the Vector Institute showcases a chemical language model used in a biological context. The authors develop a set of three chemical language models that:
Converted amino acid sequences to chemical sequence representations (SELFIES), and then generated an entire new dataset of proteins based of the linear sequence representations of proteins
Was trained on existing proteins modified with random side chain modifications, creating unnatural amino acids. The model was able to generate new proteins entirely made from unnatural amino acids.
Was trained on antibody-drug conjugates, which underwent the same conversion to linear sequence representations, to then generate entire new ADCs with novel “warheads” (small molecules).
Specialized astrocytes mediate glutamatergic gliotransmission in the CNS [de Ceglia et al., Nature, Sep 2023]
Why it matters: Glial cells have historically been thought of as simply a support cell in the central nervous system. New research has discovered a novel astrocyte subtype that is capable of rapid release of neurotransmitters and directly modulating neuronal circuits involved in memory and movement. New research published in Nature last week discovered a new cell type in the brain that is a sort of hybrid between neurons and astrocytes. These specialized astrocytes are capable of releasing glutamate, the primary excitatory neurotransmitter. The authors profiled astrocyte subtypes using single-cell RNA-seq and identified a novel subtype that expresses proteins involved in synaptic release of glutamate such as VGLUT. These glutamatergic astrocytes release glutamate on a time-scale rapid enough to modulate neuronal circuits. For example, the authors showed that astrocytes modulated activity in the nigrostriatal pathway, a dopaminergic circuit involved in voluntary movement that is dysfunctional in Parkinson’s disease, such that astrocyte VGLUT-mediated signaling exerted an inhibitory influence over dopaminergic neurons. The therapeutic relevance of this newly identified cell type is extensive, such as targeting this cell type to inhibit deep brain structures in Parkinson’s (similar to deep brain stimulation) or to oppose neuronal hyperexcitation during seizures.
What we listened to
Notable Deals
Verge Genomics teams up with AstraZeneca in drug discovery deal.
Otsuka pens AAV deal with Shape Therapeutics for eye gene therapies.
Seagen partners with Nurix to create new kind of cancer drug.
UK biotech (Apollo Therapeutics) brings in $226M Series C.
Novartis stops work on gene therapy acquired in Gyroscope deal.
Acelyrin's IL-17 inhibitor fails to hit primary endpoint in key study after splashy IPO.
In case you missed it
Curie Bio dropped their must-read Founder Guide!
Field Trip
Did we miss anything? Would you like to contribute to Decoding Bio by writing a guest post? Drop us a note here or chat with us on Twitter: @ameekapadia @ketanyerneni @morgancheatham @pablolubroth @patricksmalone