BioByte 047: multimodal AI for healthcare, tolerogenic vaccines, deciphering the lipidome, missense variant prediction, circRNAs in neurological disease, predicting TCR interactions
Welcome to Decoding Bio, a writing collective focused on the latest scientific advancements, news, and people building at the intersection of tech x bio. If you’d like to connect or collaborate, please shoot us a note here or chat with us on Twitter: @ameekapadia @ketanyerneni @morgancheatham @pablolubroth @patricksmalone. Happy decoding!
Sprinting down the streets of Manhattan? Negotiating treatises at the UN General Assembly? Make sure you’re prepared for your meetings with global leaders with the following:
An overview of the Biden-Harris Administration’s ARPA-H’s Biomedical Data Fabric Toolbox
Two biotech IPOs that breathe life into the market
Using glycosylated autoantigens to produce tolerogenic vaccines and treat autoimmune disease
A foundation model, RETFound, which succesfully diagnoses disease from retinal images
Adapting AlphaFold into AlphaMissense, which predicts the pathogenicity of missense variants
circular RNAs are widespread in neuropsychiatric disease and are linked to cell identity
TAPIR: a new language model for predicting TCR/target interactions
What we read
Blogs
ARPA-H Biomedical Data Fabric Toolbox [ARPA-H, September 2023]
The Biden-Harris Administration has introduced the ARPA-H Biomedical Data Fabric Toolbox, a project aimed at improving the accessibility of biomedical research data to advance healthcare innovations. This initiative will unify research data from various health fields, making it easier for scientists to discover and share medical insights. Led by the Advanced Research Projects Agency for Health (ARPA-H), in collaboration with other Department of Health and Human Services (HHS) agencies, this project aligns with the President's Unity Agenda and the Biden Cancer Moonshot, modernizing medical research capabilities and enhancing health outcomes. The goal is to create a comprehensive search engine for various data types, enabling quicker and more precise insights into health-related questions. Initial partners include agencies like the National Cancer Institute, National Center for Advancing Translational Sciences, and others. The project will encourage computer science and biomedical research communities to develop new search capabilities while safeguarding patient privacy and enhancing data representation. ARPA-H is committed to advancing health data science and accelerating solutions for better patient outcomes through collaboration across HHS and the broader health ecosystem.
RayzeBio, Neumora price some of biotech’s largest IPOs this year [BiopharmaDIVE, Gwendolyn Wu, September 2023]
Neumora and RayzeBio, two biotech companies developing new medicines for cancer and brain diseases, held large initial public offerings (IPOs) this past week. Neumora sold 14.7 million shares at $17 each, raising $250 million. RayzeBio sold 17.2 million shares at $18 each, raising $311 million. This is the second largest biotech IPO of 2023 so far. The successful offerings may signal a return of investor demand for biotech stocks after a long period of weak IPO performance. Strong investor backing, experienced leadership teams, and advanced drug pipelines contributed to the success of these IPOs.
In total, only 15 biotech IPOs have been completed in 2023, versus 183 in 2020-2021. Most biotech IPOs this year raised under $100 million. Experts say IPO activity will remain slow compared to past years, but there are positive signs like the large sums raised by Neumora and RayzeBio. More biotech IPOs may shift to reverse mergers with existing public companies. Overall, the biotech IPO market shows early signs of rebound but is not yet back to previous high levels of new offerings. Companies with strong financial backing and drug candidates in late stage testing remain best positioned to complete IPOs.
As artificial intelligence goes multimodal, medical applications multiply [Eric Topol, Science, 2023]
We’ve been reviewing the ever growing use cases and tribulations of LLMs in healthcare for the past year. Much of the attention of these early applications have been placed on their ability to pass the US medical licensing exam, provide answers to medical questions and reduce bureaucratic burden of clinicians which use a single mode of data; text.
There have been many attempts at integrating a few layers of data, such as EHRs and genomics, but have lacked the breadth of what can actually be analyzed. As Eric puts it “that represents a considerable ongoing challenge to actualize the extraordinary potential of multimodal AI in medicine.”
Unlocking true multimodal AI would open up the opportunities that could significantly improve healthcare cost burden, patient prognosis and patient experience. For instance a virtual health assistant could provide frequent feedback to patients at risk of chronic diseases or allow for true remote monitoring with continuous vital-sign capture that is equivalent to an intensive care unit.
However, there are many barriers beyond the analytical/modality integration. LLMs are overconfident, they confabulate, have embedded biases, there is resistance from the medical practice to change and open questions about what is necessary for regulatory approval.
An Exciting New Approach to Autoimmune Diseases [Eric Topol, Ground Truth, September 2023]
Eric Topol started this essay off with quite the hook:
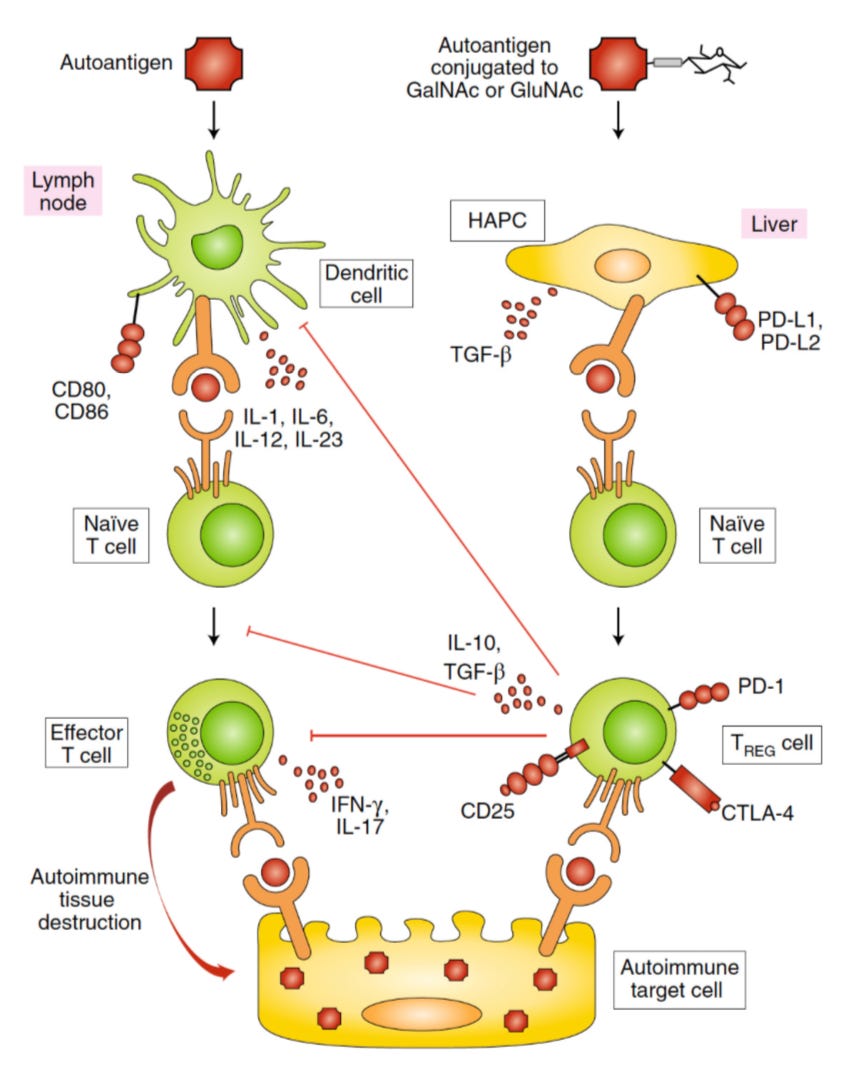
The type of vaccine he’s referring to is known as a tolerogenic vaccine that essentially induces immune tolerance (not attacking self) while also not shutting the rest of the immune system down. This is a complex problem—riddled with challenges in making sure you’re not stimulating the whole immune system, knowing the right autoantigens to target in the first place, inducing the right degree of tolerance, etc. The University of Chicago research that Topol covers in this piece specifically focuses on using the liver to induce tolerance. Why the liver? The liver is a unique organ when it comes to immune tolerance because it is considered tolerant and sees a flux of immune cells while serving as a source of many antigen-presenting cells via hepatocytes. Topol covers research from the Hubbell lab on how you can take advantage of the liver’s immune tolerant systems and modify autoantigens via glycosylation (adding a sugar) to modulate disease. In simpler terms, the liver is involved in telling the body what cells to leave alone as they are getting processed to avoid an autoimmune reaction to naturally dying cells but by glycosylating an autoantigen with a molecule the liver recognizes, you’re tricking the process. This method, if it works, can be used in autoimmune disease treatment and prevention.
Academic papers
A foundation model for generalizable disease detection from retinal images [Zhou et al, Nature Medicine, September 2023]
Why it matters: RETFound demonstrates the potential of SSL and foundation models to alleviate key barriers in medical AI, including need for large labeled datasets and limited model generalizability. By efficiently leveraging abundant unlabeled medical images, RETFound can enable rapid development of high-performing AI across diverse tasks. This research has major implications for ophthalmology and other specialties. RETFound also highlights retinal imaging as a valuable window into systemic health. The ability to predict heart disease and other conditions from retinal scans could enable risk stratification and earlier intervention.The paper introduces RETFound, a new foundation model for retinal image analysis based on self-supervised learning (SSL). RETFound was trained on a large dataset of over 1.6 million unlabeled retinal images, including both color fundus photos and optical coherence tomography scans. After pretraining with SSL, RETFound can be efficiently fine-tuned for a wide range of downstream tasks using small labeled datasets.
The researchers evaluated RETFound on tasks including diagnosis of retinal diseases like diabetic retinopathy, prognosis of conditions like wet AMD, and prediction of cardiovascular diseases from retinal images. Across all tasks, RETFound achieved significantly higher performance compared to other models, even those pretrained on ImageNet. It also required less labeled data to adapt to new tasks. Qualitative analyses showed RETFound learns to identify relevant anatomical structures for each disease.
Dynamic lipidome alterations associated with human health, disease and ageing [Hornburg et al., Nature Metabolism, September 2023]
Why it matters: This extensive, longitudinal lipidomic profiling study provides new insights into the distinct roles of lipid species and subclasses in human health and disease. The data serve as a valuable resource for exploring lipid involvement in metabolism, inflammation, immune function, and aging. Defining lipid signatures of early disease onset could enable new monitoring strategies. The study also suggests potential areas for therapeutic interventions, such as dietary supplementation to restore beneficial lipid levels. Overall, this work demonstrates the power of deep lipidomics to elucidate the complex lipid-phenotype connections important for precision health.The researchers performed comprehensive lipidomic profiling on over 1500 plasma samples from 112 participants followed longitudinally for up to 9 years. They quantified over 800 lipid species across 16 subclasses. Many lipids showed participant-specific signatures that were stable over time.
Lipid modules correlated with clinical markers of metabolic health, inflammation, and diabetes. Key lipid changes were observed in insulin resistance, during respiratory viral infections, and with aging. For example, triglycerides and diacylglycerols increased in insulin resistance while ether-linked phosphatidylethanolamines decreased. Distinct lipid species changed rapidly during viral infection and recovered at different rates. Aging was associated with increased levels of ceramides, sphingomyelins, and cholesterol esters, but decreases in polyunsaturated fatty acids.
Furthermore, the study found over 1200 associations between specific lipids and cytokines/chemokines. This provides insights into the diverse immunoregulatory roles of lipids.
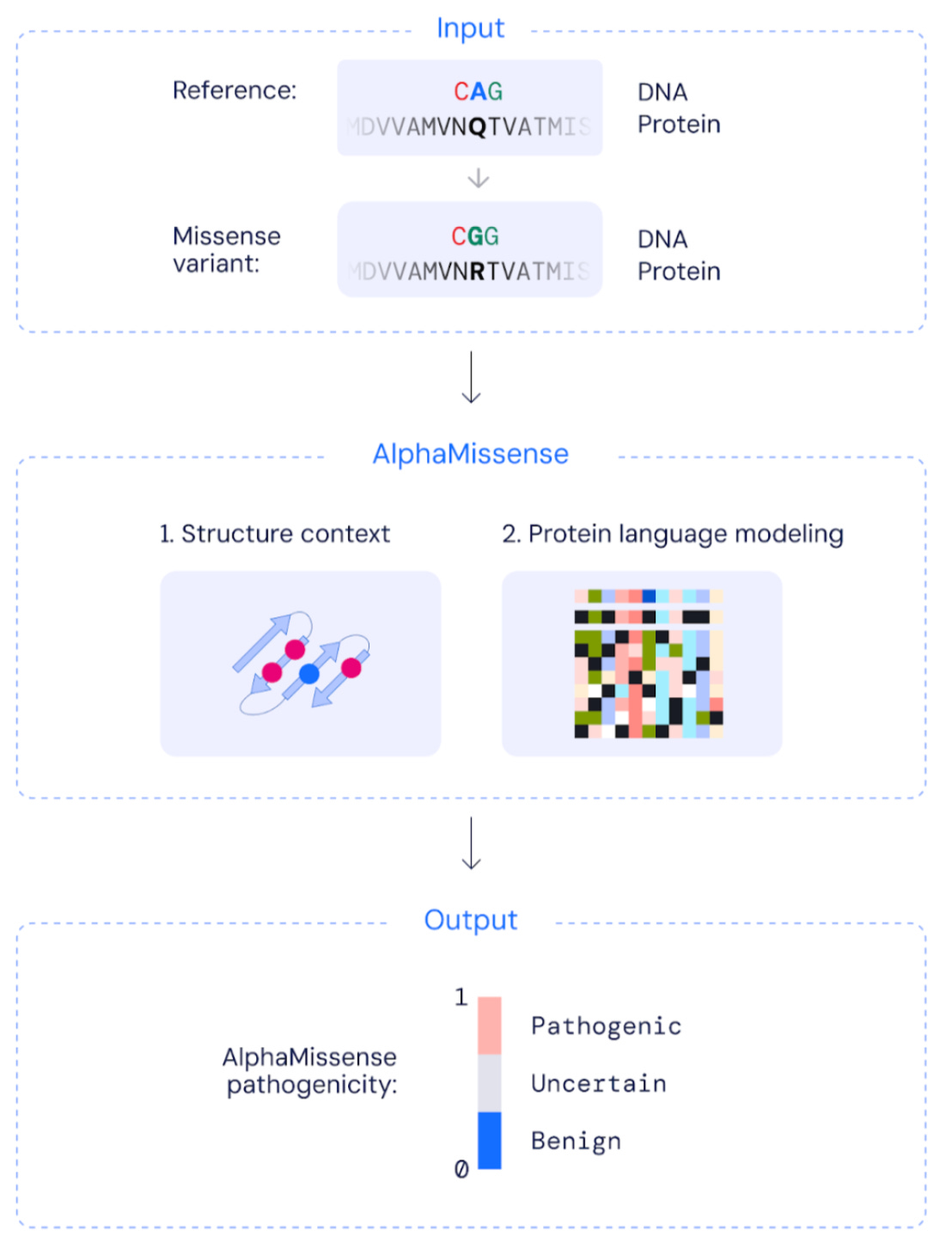
Accurate proteome-wide missense variant effect prediction with AlphaMissense [Cheng et al., Science, Sep 2023]
Why it matters: The average person is carrying 9k+ missense mutations. Most are benign, but others can disrupt protein function or cause disease. To date, only 0.1% of the 71 million possible missense variants have been classified as benign or pathogenic. This week in Science, a Deepmind team adapts AlphaFold and protein language models into a new AI system called AlphaMissense, and successfully predicts the pathogenicity of missense variants.The Deepmind team has released a new model called AlphaMissense, which is capable of predicting the pathogenicity of missense variants (a single nucleotide substitution that changes the coded amino acid, potentially changing protein function and causing disease). AlphaMissense combines several pre-trained models including AlphaFold for protein structure prediction and protein language for sequence modeling, and fine-tunes on labels distinguishing variants seen in human and closely related primate populations. Variants commonly seen are treated as benign, and variants never seen are treated as pathogenic. The method achieves SOTA performance on several benchmarks, and was used to predict the effects of 71 million missense mutations which are now published in an online database.
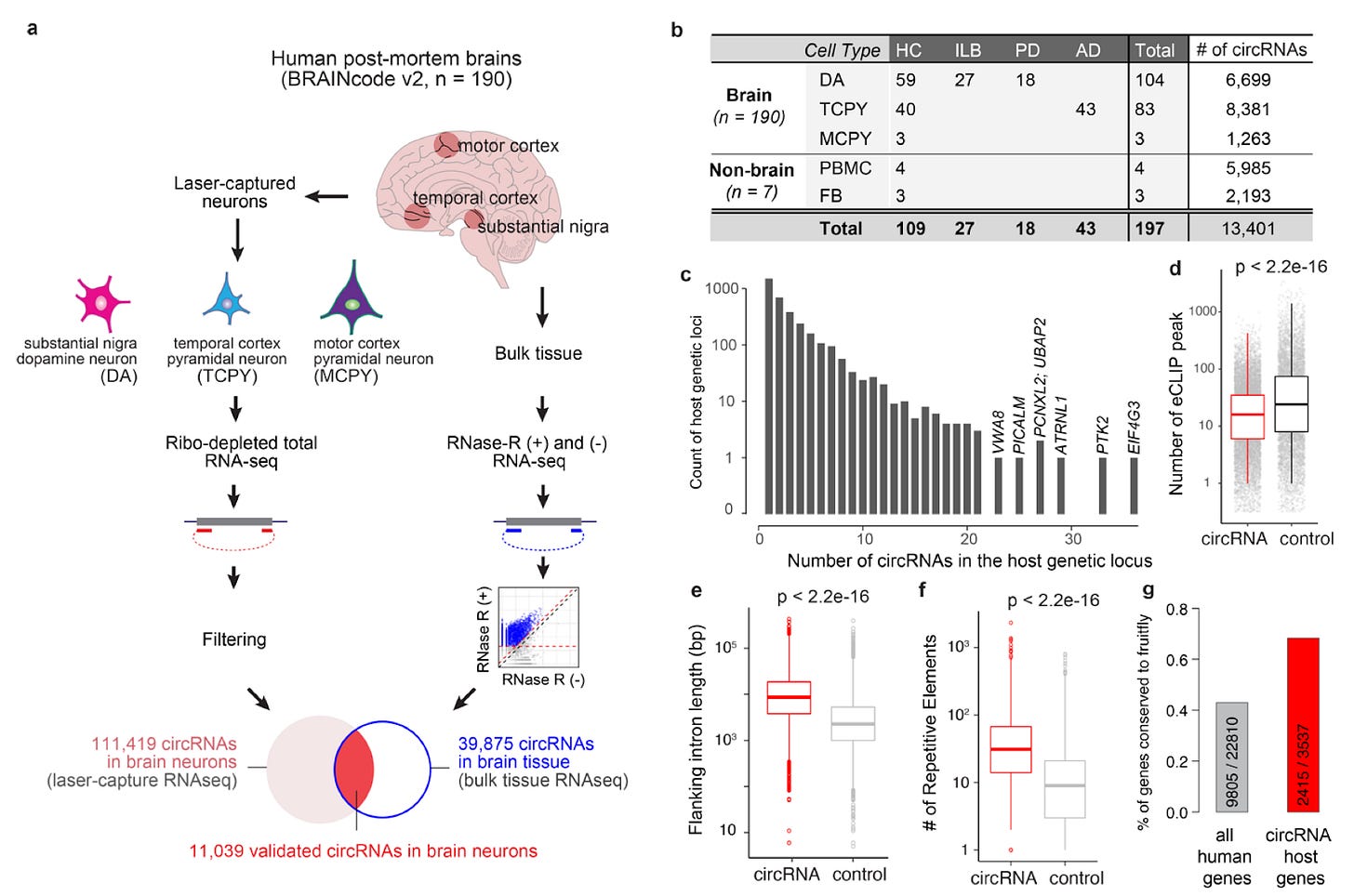
Circular RNAs in the human brain are tailored to neuron identity and neuropsychiatric disease [Dong et al., bioRxiv, 2023]
Why it matters: Circular RNAs (circRNAs) were largely thought to be waste products from mRNA splicing, with little to no function in disease. However, work within the past decade has begun to completely overhaul our understanding of circRNAs - that they have significant implications across transcriptional and translational dynamics. In this paper, Dong et al demonstrate that circRNAs in the human brain are bespoke according to cell type, and are enriched in neurodegenerative disease, underscoring their role in synaptic dynamics.Here, the team laser-captured neurons and non-neuronal cells from postmortem human brain samples before using ultra-deep total RNA-seq to probe RNA sequence diversity. Notably, 61% of synaptic circRNAs were associated with 20 neuropsychiatric diseases. Additionally, they found several cell-type specific circular RNAs in dopaminergic and pyramidal neurons; surprisingly, these were more likely to define neuronal identity as compared to linear RNAs. Focusing on disease, they also found that several genes implicated in Parkinson’s and Alzheimer’s produced circular RNAs. This link across disease and cell-type was reinforced when they found that disease-linked circRNAs also demonstrated cell-type bias. For example, addiction-associated genes mainly expressed circRNAs in dopaminergic neurons (implicated in addiction), autism genes expressed circRNAs in pyramidal neurons, and Parkinson’s Disease associated genes produced circRNAs in neurons and nonneuronal cells. Thus, the authors speculated that circRNAs may play a role in assembling cell-type specific synapses and dysregulation may contribute to synaptic-driven disease. Although significant work still remains to understand the function of said circRNAs, this paper sheds light on the importance of this class of non-coding RNAs in neurological function and disease states
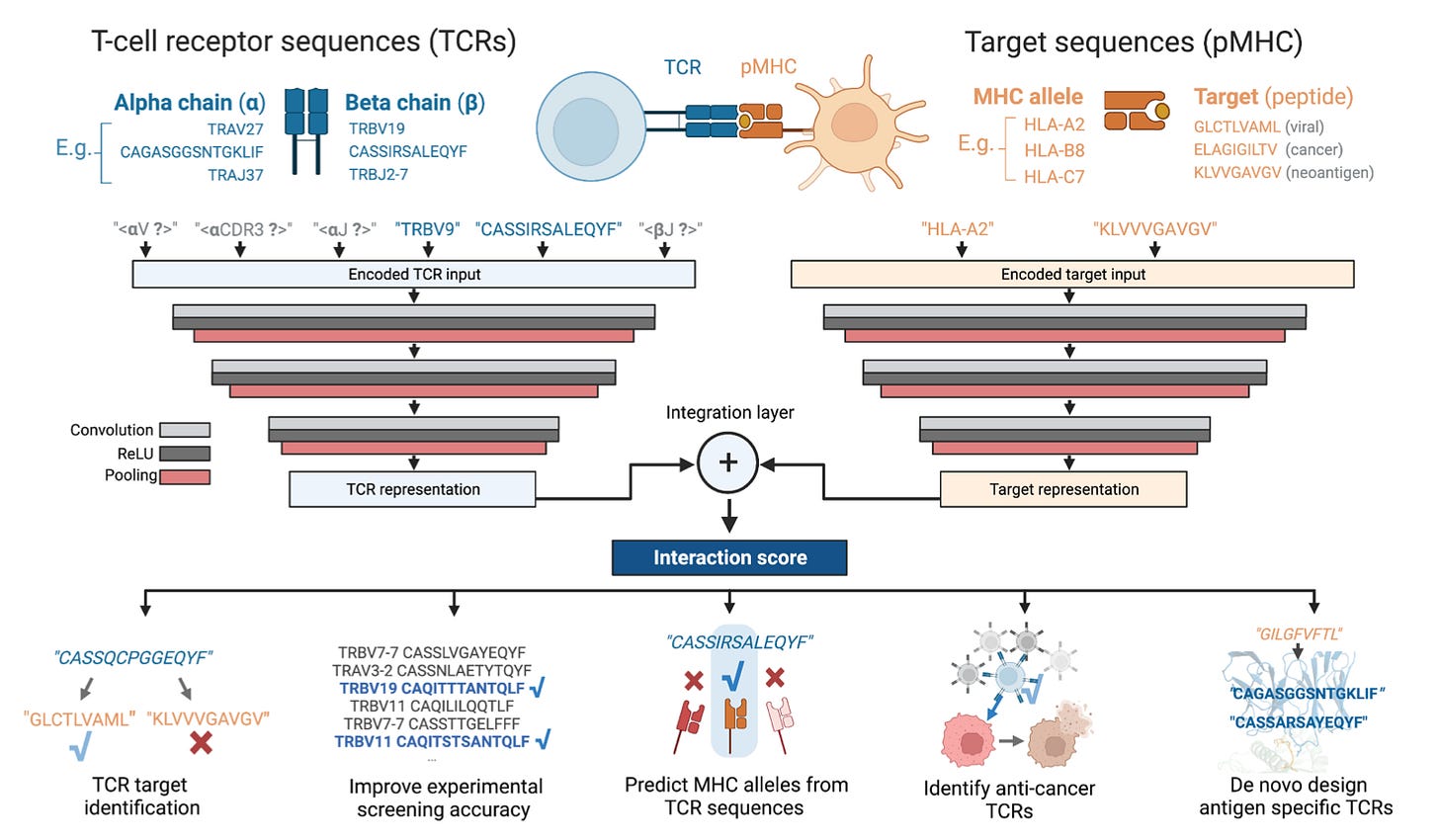
TAPIR: a T-cell receptor language model for predicting rare and novel targets [BioRxiv, Fast et al., September, 2023]
Why it matters: Linking TCRs (T-cell receptors) with their targets holds a lot of promise for uncovering links in immunology and treating associated diseases. This study presents a language model for predicting such TCR/target interactions which limited groups have been able to comprehensively do. The authors have created TAPIR—T-cell receptor and peptide interaction recognizer by using convolutional neural network encoders to read in TCR and target sequences and using that to train a model to learn themes across their interactivity. TAPIR notably can train on paired and unpaired TCRs and can predict interactions against novel targets not exposed in training tasks—this breakthrough in particular has not yet been done to our knowledge. The authors demonstrated the inverse, that TAPIR can also design TCR sequences if given a target of interest. This model lays the foundation for several useful tasks in medicine including TCR prediction for diagnostics, vaccine design, specifically cancer vaccine design, and even some cell therapies. The group launched startup VCreate to commercialize these efforts last year.
What we listened to
Notable Deals
Backed by Illumina, Broken String Biosciences raises $15M to find off-target gene edits
RNA editing startup launches with $30M based on Stanford and University of Tübingen research
With covalent meds abuzz, RA Capital and Novartis lead $56M seed round for new startup
In case you missed it
What we liked on Twitter
Events
September 21st – Grand Discoveries 2.0: A Hitchhiker’s Guide to TechBio and the future of health, Cambridge (UK)
Registration at https://lu.ma/oxubz1sl!
Field Trip
Did we miss anything? Would you like to contribute to Decoding Bio by writing a guest post? Drop us a note here or chat with us on Twitter: @ameekapadia @ketanyerneni @morgancheatham @pablolubroth @patricksmalone










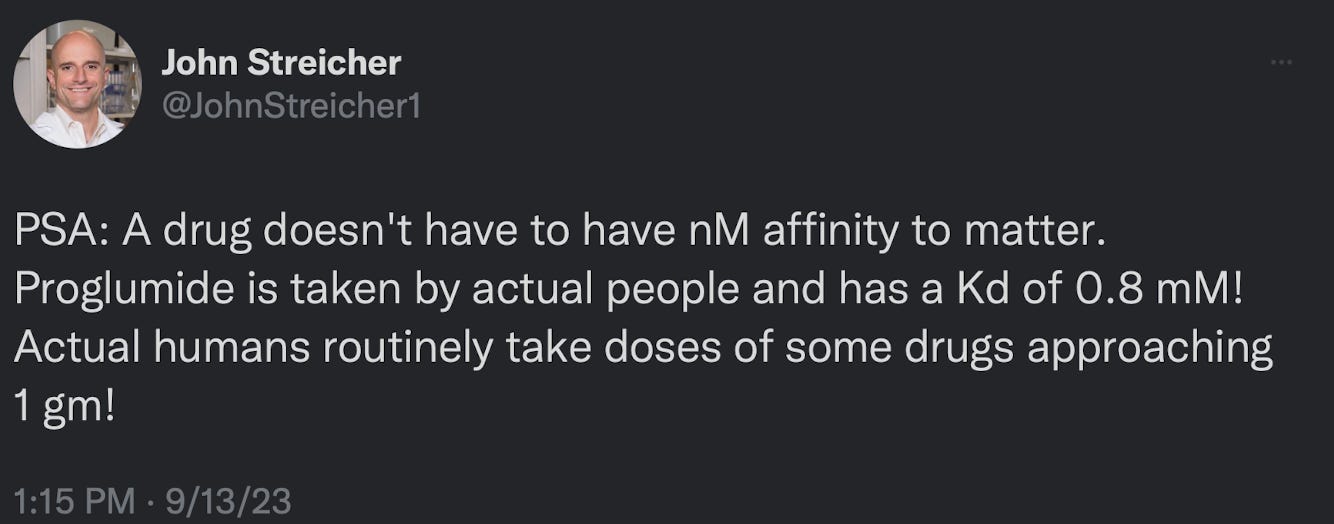

Appreciate the podcast recommendations!