BioByte 049: atomwise focuses on pipeline, AI gets the american nobel prize, predicting transcriptional outcomes, nanopore protein sequencing detects all amino acids, deep learning for cryo-EM
Welcome to Decoding Bio, a writing collective focused on the latest scientific advancements, news, and people building at the intersection of tech x bio. If you’d like to connect or collaborate, please shoot us a note here or chat with us on Twitter: @ameekapadia @ketanyerneni @morgancheatham @pablolubroth @patricksmalone. Happy decoding!
We have two new updates today:
Zahra Khwaja from Charm Therapeutics has masterfully curated BioByte’s Notable Deals section to include brief analyses. Check it out below.
Our very own Pablo Lubroth will be climbing Mt. Kilimanjaro alongside other biotech founders, scientists, investors and operators as part of the Timmerman Traverse team in February. He’ll be raising $50k for the Damon Runyon Cancer Research Foundation, contributing to a total of $1m raised by the team.
The DRCRF funds high-risk, high-reward cancer research. They identify and enable young scientists who are bold enough to go where others haven’t. Incredible scientific talent goes unfunded because they don’t fit the norm and perhaps think so far outside the box, that it makes them ineligible for existing grant funding. This is particularly aligned with our mission at Decoding Bio — we don't need to tell you that a good chunk of important science flies under the radar (cc Katalin Karikó).
Damon Runyon has set out to change that. The Foundation’s scientists have developed tools like CRISPR-Cas9, discovered the link between HPV and oral cancer, and account for 13 Nobel laureates in their roster. It truly generates outsized outcomes per dollar spent.
If you are in a position where you could donate, we would be so grateful. It’ll have a meaningful impact on cancer patients and the Foundation. We'll be sure to keep you posted on Pablo and the rest of the Timmerman Traverse team's journey later this year.
What we read
Blogs
'Faster isn't better' — Atomwise moves from partnerships to pipeline with TYK2 drug [Andrew Dunn, Endpoints, 2023]
In this interview with Abraham Heifets, CEO and Founder of Atomwise, he talks about the pivot from high volume pharma partnerships to developing an internal pipeline. Atomwise has signed more than 750 partnerships; mainly companies asking for binders for a target protein. But none of those molecules have made it to the clinic. The company has now decided to refocus into its internal pipeline, starting with TYK2 which has yielded lucrative acquisitions in the past.
“Faster isn’t better,” Heifets said. “Faster to failure is not success. Cheaper to failure is not success.”
AI is no silver bullet to Eroom’s law. The first AI drug discovery companies including Atomwise, BenevolentAI and Excientia the the use of new computational techniques without an insight as to why this would increase the chances of approval of drug, it is not guaranteed that they will produce successful drug candidates.
How to represent a protein sequence [Liam Bai, September 2023]
One of the most important aspects of machine learning, especially in computational biology, is how input data is transformed into vector representations, a process called representation learning. In this essay, Liam Bai provides a primer on representation learning for proteins by walking through an example of a BERT-like encoder model (bidirectional encoder representations from transformer, a common NLP algorithm developed at Google) applied to protein sequencing modeling. A common approach in protein language models, borrowed from the NLP field, is masked language modeling, a form of self-supervised learning in which a model is trained to predict hidden tokens (e.g., words, or amino acids). The amazing thing about these models is that they result in vector representations that capture information about protein structure (and other interesting properties) without being explicitly trained to do so. For example, by inspecting the attention maps of the trained models (a visualization of how a transformer attends to different parts of an input sequence, commonly used to understand how the model processes the input data), it is observed that amino acid pairs with high attention scores are more often in 3D contact in the folded structure.
A New Precedent—AI Gets the “American Nobel” Prize in Medicine [Eric Topol, Ground Truths, 2023]
Katalin Kariko and Drew Weissman were awarded the Nobel prize in medicine earlier this week for their important mRNA work that led to the creation of the COVID-19 vaccine. Another prestigious award in medicine, one that is often used to predict future Nobel laureates, was awarded to Demis Hassabis and John Jumper for their work with AlphaFold. This award, the Albert Lasker Foundation Basic Medical Research Award, is lauded as the highest medical science award in the US aka “the American Nobels”. This is an interesting choice for an award recipient because though the award is officially awarded to Hassabis and Jumper, it is for the program itself, which is artificial intelligence. And because AlphaFold utilizes transformer models, we don’t actually know how it works, as Topol writes, there’s a “combination of human ingenuity and the black box of a transformer model”. This presents an interesting point and perhaps foreshadowing for future awards and also just the way we view scientific advancement—The rest of the post provides a great overview of AlphaFold, its current uses, limitations, and expansion cases.
Six imperatives for building AI-first companies [Morgan Cheatham, TechCrunch]
Decoding Bio contributor, Morgan Cheatham, posted a byline in TechCrunch last week detailing the “Six Imperatives for building AI-first companies,” outlining key distinctions between AI-first and AI-enabled companies. The piece draws upon examples from healthcare and life sciences spanning AI for radiology, computer vision biomarkers, and clinical informatics.
Definitionally, the article describes AI-first companies as those that are in the business of advancing AI as a science, whereas AI-enabled companies are implementation and distribution machines. The two company phenotypes establish moats at different layers – AI-first companies innovate just above silicon, while AI-enabled companies create enterprise value at the application-level.
Create and Sustain an Undeniable Data Advantage: AI-first companies prioritize acquiring and creating unique, high-quality data sets. This includes designer data sets tailored to specific tasks and domains. It's not just about using existing data but also generating novel data assets through experimentation and customer input.
Recruit and Empower AI Scientists: AI-first companies assemble multidisciplinary teams that combine AI research expertise with domain knowledge, such as healthcare professionals collaborating with AI researchers. The organizational structure reflects a strong commitment to AI, often with a Chief Scientific Officer overseeing AI research.
Support a Flexible AI Stack: AI-first companies avoid rigid decisions about their AI stack, focusing on modular approaches that incorporate publicly available models where appropriate. This flexibility allows them to innovate lower in the AI stack, leading to greater product versatility.
Establish Distribution Moats: Both AI-first and AI-enabled companies need distribution strategies, but AI-first companies benefit from technical and distribution moats. These moats increase the chances of success and enable integration with or displacement of incumbent technology providers.
Center Safety and Ethics in Model Development: AI-first companies prioritize safety and ethics in their foundational AI research. They implement continuous monitoring, fail-safes, human intervention overrides, and ongoing validation against real-world data. User training is critical to communicate AI limitations.
Earn and Maintain Trust: Trust is paramount for AI-first companies, built through reliability, accuracy, and a genuine commitment to solving problems with empathy. By following these imperatives, AI-first companies can earn and maintain the trust of users, customers, and their industry.
Academic papers
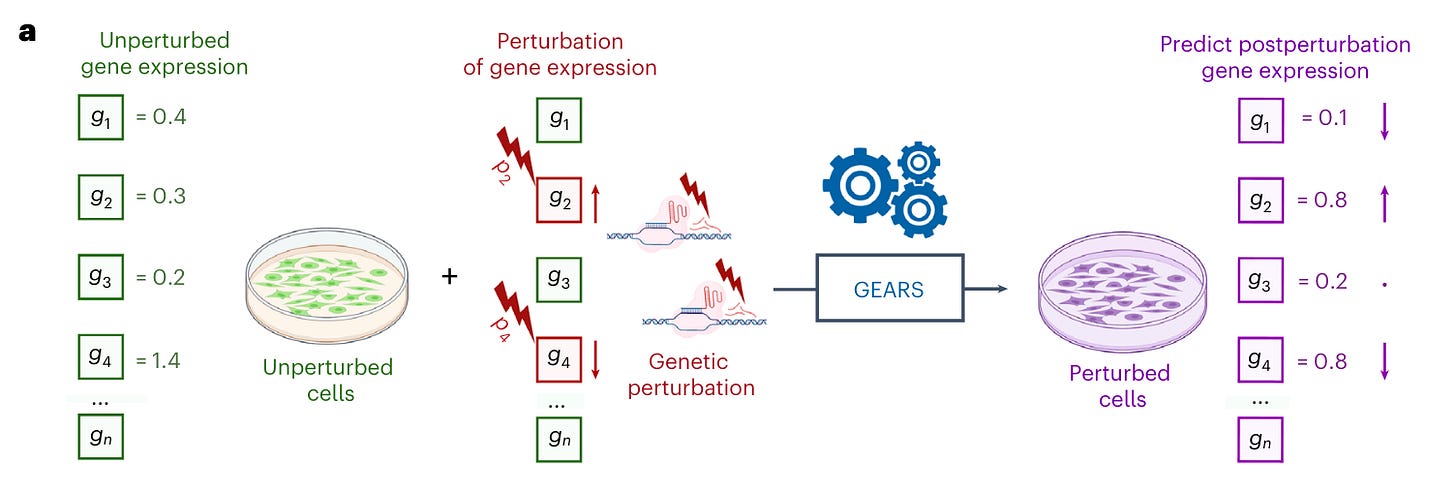
Predicting transcriptional outcomes of novel multigene perturbations with GEARS [Roohani et al., Nature Biotech, 2023]
Why it matters: understanding phenotypic responses based on genetic perturbation is critical for the validation of new drug targets, identifying genetic interactions in cancer, developing drug combinations and many other applications. The authors present a model that can predict transcriptional outcomes for gene perturbations for which there is no experimental data.These Stanford researchers developed GEARS (graph-enhanced gene activation and repression simulator), a method which integrates deep learning with a knowledge graph of gene-gene to predict transcriptional responses using scRNASeq data from perturbational screens. Importantly, GEARS can predict outcomes for gene perturbations that were not experimentally tested. This is is contrast to current methods:
For single-gene perturbation prediction, the main common method relies on the inference of transcriptional relationships between genes in the form of a gene regulatory network. This is limited by inference accuracy and the incompleteness of the networks.
Current predictive models miss on synergistic effects of genes, given they can only linearly combine the effects of individual perturbations
Recent work using deep neural networks can skip the inference step and map genetic relationships into latent space, but each gene in the combination has to be experimentally perturbed before the effect of the perturbation in the combination can be predicted.
Given that GEARS can infer a broader range of perturbation outcomes using the same experimental data, it can be useful as a supplementary method to experimental perturbation experiments. It can also be used to direct and design the experiments that will yield the most informative experiments.
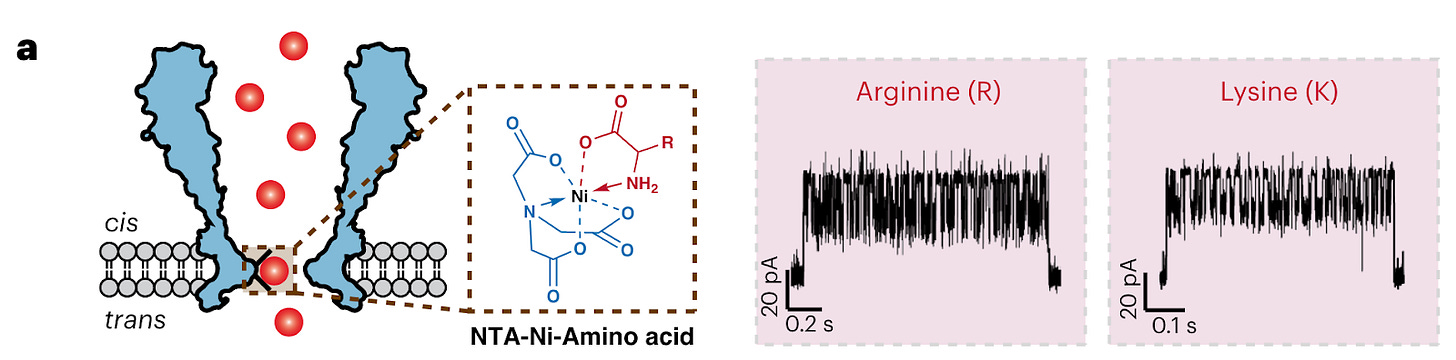
Unambiguous discrimination of all 20 proteinogenic amino acids and their modifications by nanopore [Wang et al., Nature Methods, September 2023]
Why it matters: In Nature Methods this week, a new proof-of-concept demonstration of nanopore-based identification of amino acids and associated post-translational modifications. The impact of a nanopore-based protein sequencer would be significant, including more sensitive detection of proteins, improved characterization of post-translational modifications, and portable protein sequencing for applications such as point-of-care diagnostics. Nanopore protein sequencing is an emerging technique that allows direct reading of amino acid sequences by monitoring changes in electrical current as a protein translocates through a nanopore. Several companies use this technology for other biological molecules, such as Oxford Nanopore, which develops and commercializes nanopore-based DNA and RNA sequencing technology. The advantage of nanopores is that they permit single-molecule characterization, which is important for sensitivity (many proteins exist in low concentrations) and detecting post-translational modifications (PTMs).
All 20 amino acids and some PTMs could be discriminated through a nanopore engineered with a nickel ion on the inside of the channel (image below, left). As each amino acid passes through the channel, it changes the potential of a voltage applied across the membrane (image below, right). A machine learning model was trained on the current traces and achieved 98.6% accuracy in classifying amino acids.
The impact of a nanopore-based technology for protein sequencing would be significant. Existing methods for protein sequencing such as mass spec are indirect, and therefore a direct method like nanopores would provide a more accurate view of protein sequence. Nanopore-based methods will also more accurately characterize PTMs, largely missed by existing methods (and protein-folding algorithms like AlphaFold), which have an important influence on protein structure. Finally, nanopore-based sequencers are more portable, and therefore unlock applications like point-of-care diagnostics.
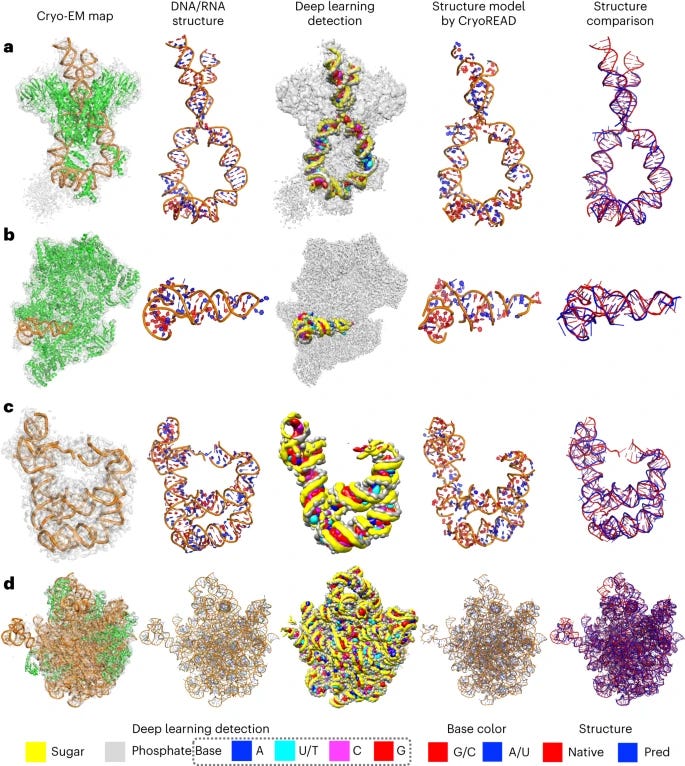
CryoREAD: de novo structure modeling for nucleic acids in cryo-EM maps using deep learning [Wang et al, Nature Methods, 2023]
Why it matters: cryogenic electron microscopy (cryo-EM) is a great tool for elucidating structure of proteins and some nucleic acid/protein interactions. However, it is difficult to use such tools for understanding structure at a resolution high enough to look at DNA and RNA structure because cryo-EM is limited by resolution and mostly geared towards studying proteins. There are few computational models for modeling the structure of nucleic acids. The authors of this paper developed CryoREAD, an automated de novo nucleic acid atomic structure modeling method based on deep learning. The system works by identifying phosphate, sugar, and base positions fundamental to nucleic acid structures in a cryo-EM map with deep learning and then modeling that into a 3-D structure. The sugars are connected to form a backbone and the nucleic acid sequence is projected onto that. The system was tested at the 2-5 Å resolution, medium-scale resolution. They found that 85.7% of atoms were placed correctly within 5Å and 52.5% of nucleotides were correctly identified. The authors also tested their CryoREAD model against existing software that is mainly used for protein reads but is often adapted for understanding nucleic acids. While limitations of nucleotide sequence prediction need to be addressed, this work presents a foundation for DNA/RNA structure modeling without manual human input. This can help fill a fundamental tool gap in structural biology and modeling.
DisP-seq reveals the genome-wide functional organization of DNA-associated disordered proteins [Xing et al., Nature Biotechnology, 2023]
Why it matters: Many proteins (such as transcription factors) contain Intrinsically disordered regions (IDRs) – areas which lack a fixed three-dimensional structure – are known to play an important role in gene regulation. However, we still lack a sophisticated understanding of their distribution and impact across the genome. In this paper, the authors develop DIsP-seq (disordered protein precipitation followed by DNA sequencing), allowing them to decipher the distribution and function of IDRs.To date, technologies for probing DNA-binding IDRs use antibodies to precipitate proteins; this generally requires the addition of an engineered tag, or working with those that are well studied and have known binders. IDRs are fluid and transition across several conformations - making their functional characterization challenging. Here, the authors use biotinylated isoxazole (binds to proteins with low-complexity regions - regions of biased composition, containing simple sequence repeats) to selectively precipitate IDR-containing proteins bound to DNA. They then treat the samples with nuclease to fragment unprotected DNA, and also use proteinase K, and RNase before massively parallel sequencing and mapping the reads to the genome. This readily uncovers the distribution of IDRs across the genome, which can then help identify binding motifs and enriched regions. The authors used DisP-Seq to probe the oncogenic fusion protein EWS-FLI1 (in which the IDR from ESWR1 is fused to the DNA-binding domain of the transcription factor FLI1) in Ewing Sarcoma, and demonstrate that cell-type specific spatial organization of DNA-binding IDR-containing proteins can significantly impact pathologic gene regulatory dynamics.
Biomedical publishing: Past historic, present continuous, future conditional [PLOS Biology, Sever, October 2023].
Why it matters: This paper provides an insightful analysis of the history of scientific publishing and peer review, making the case that internet technologies enable us to rethink outdated publishing models and build a more open, transparent and equitable scientific communication system. The ideas presented could help address systemic issues in academia relating to evaluation and incentives. However, achieving such a vision would require concerted efforts and buy-in across various stakeholders.This paper provides a historical overview of the evolution of scientific publishing and peer review over the past 350 years. It highlights how peer review was not routinely used until the late 20th century and has become conflated as a marker of quality and impact. The paper argues that in the internet age, we have an opportunity to decouple dissemination and evaluation of research. Preprint servers now allow immediate sharing of manuscripts before formal peer review. The author advocates for a more open ecosystem in which preprints are posted first, then undergo various forms of review, curation and evaluation from multiple sources over time. This could avoid issues with the current system like the over-reliance on journal brands and impact factors as proxies for quality. Content verification and badging of articles could provide better multidimensional trust signals. The author cautions against publisher "lock-in" of content and outlines principles for building a more equitable and transparent system.
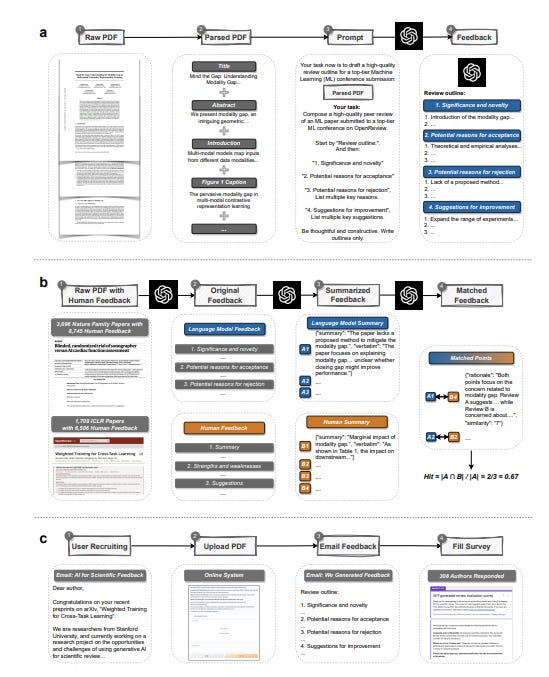
Can large language models provide useful feedback on research papers? A large-scale empirical analysis [Liang et al., arXiv, October 2023]
Why it matters: The ability for LLMs to provide helpful scientific feedback at scale could greatly benefit researchers lacking access to quality peer review. It also has the potential to assist authors in improving their work prior to journal submission. However, human expert feedback remains indispensable, and LLM-generated feedback should be considered complementary rather than a wholesale replacement. Responsible deployment of LLMs is critical to avoid undermining rigorous peer review. Overall, this rigorous empirical analysis sheds light on the promises and perils of deploying LLMs to support the scientific process.This paper presents a large-scale analysis evaluating the ability of large language models (LLMs) like GPT-4 to generate helpful and reliable scientific feedback. The authors developed a system using GPT-4 that can take in a research paper PDF and output structured feedback on aspects like novelty, reasons for acceptance/rejection, and suggestions for improvement.
Through retrospective analysis on over 4,800 papers from top journals and conferences, the authors found that around 30-40% of feedback points raised by GPT-4 overlapped with individual human reviewers. This level of overlap is comparable to that between two human reviewers. The overlap was even higher for weaker papers. In a prospective user study with over 300 researchers, most found the LLM-generated feedback helpful and potentially beneficial for authors.
However, the study also uncovered limitations of LLMs for scientific feedback. The feedback tends to focus on certain aspects more heavily compared to human reviewers, and also struggles to provide concrete and actionable suggestions at times.
What we listened to
Notable Deals
M&A: Lilly Acquires Point Biopharma for $1.4B
Eli Lilly: Indianapolis-based pharmaceutical giant ($LLY , market cap: $504B)
Point BioPharma: Next-generation radioligand therapies for cancer treatment ($PNT)
Deal value and terms: Lilly is paying $1.4B ($12.50 per share) in an all cash deal that is 87% higher than Point’s closing price before the deal was announced (October 2nd)
Rationale: An attractive emerging therapeutic:
Biology: Radioligand therapies deliver radiation directly to cancer cells only, minimising contact with the healthy surrounding tissue. This ensures a more potent treatment that avoids many of the terrible side effects seen with standard radiation therapy.
Market: Novartis has demonstrated commercial blockbuster potential with its two radioligand therapies Pluvicto (PSMA+ Prostate Cancer) and Lutathera (Somatostatin+ gut tumours) which bring in around $1.6B / year combined.
High barriers to organic entry: 1. Critical to have secure and plentiful radio-isotope suppliers: Point has built multiple strong Lutetium and Actinium supplier relationships across the globe. 2. Complex supply chain and manufacturing: Point has developed a fortified supply chain, with built-in redundancies to ensure resilience. Point has built CORE, a 180,000 sq ft manufacturing facility in Indianapolis
Point Biopharma’s pipeline: Late stage: although phase III assets here are derisked and likely to achieve clinical success (similar payload and target to Novartis’ approved therapies) they are out-licensed, but Point has retained a significant royalty share of 15-20%. Early stage: arguably the real value driver, Point has novel targets (FAPa) in the pipeline with actinium (over Lutetium) payloads that are touted to be safer to administer by physicians and more lethal to cancer cells
Financing: Iambic (formerly Entos) raises $100M Series B
Iambic: AI-driven biotech with a platform of proprietary deep learning models for small molecule drug design
Key models: OrbNet: predicts molecular properties / energies using atomic orbitals. NeuralPlexer: diffusion generative model for protein-ligand structure prediction
Team of 60 employees split between drug discovery and AI experts
Deal value and terms: Raised $100M from Nvidia, Ascenta Capital, Abingworth, Illumina Ventures, Gradiant Corporation as well as original backers such as Orbimed. (Launched initially with $53M)
Rationale: healthy pipeline of disclosed candidates:
IAM-H1 HER2 TKI (IND-enabling): HER2 landscape is extremely crowded, and Iambic are also developing an orthosteric binder potentially limiting true selectivity potential
Dual CDK2/4 inhibitor (IND-enabling): Potential first-in-class and very compelling target. CDK4/6 inhibitors have seen blockbuster success in the ER+ breast cancer market with combined market sizes of $30B+ from palbociclib, abemaciclib and ribociclib. However, CDK6 is believe to be a key culprit of adverse events in some patients. Additionally, recent data has implicated CDK2 in resistance to CDK4/6 inhibitors and targeting CDK2 re-sensitises tumours to CDK4/6 inhibitors in vitro and in mice. This provides a strong rationale for a dual CDK2/4 inhibitor.
A thought: Although the shape of the pipeline is strong, it would be interesting to evidence on whether these compounds were truly enabled by Iambic’s platform or rather by the expertise of their medicinal chemists.
Collaboration: Ginkgo Bioworks and Pfizer
Ginkgo Bioworks: A broad company specialising in a variety of tools and platforms to engineer biology for a variety of applications in drug discovery, agriculture and materials.
Pfizer: American multinational pharma giant ($PFE, market cap: $189B)
Deal value and terms: Ginkgo will receive an aggregate of $331M and royalties on three RNA-based programmes of Pfizer’s choosing. Pfizer will leverage Ginkgo’s RNA Codebase platform and Foundry to enable faster discovery of RNA-based drugs
Rationale:
Circular RNA know-how: Ginkgo recently acquired Circularis, a biotech with a novel circular RNA platform. Circular RNA is touted to offer greater stability, programmability and durability compared to traditional mRNA, making it attractive for pharma’s looking to remain competitive in RNA therapeutics.
Unique platform and data generation capability: Ginkgo has a large synthetic biology platform where they can exploit various tools to meet Pfizer’s needs. Ginkgo has a 100,000 square feet of lab space with heavy automation for efficient data generation
A thought: Circular RNA therapeutics are still a risk, this therapeutic has yet to prove itself and is more complicated to manufacture than linear mRNA. This could potentially be why the financial terms have not been broken down into upfront / milestones in the press release, perhaps the financials are significantly backended.
TL1A fever continues with Sanofi and Teva’s $1.5B pact
Teva: US/ Israeli generics company. Following acquisition of Allergan’s generics unit became from of the largest globally.
Sanofi: French multinational pharmaceutical and healthcare company headquartered in Paris, France.
Deal value and terms: Upfront: $500M. Milestone (phase III initiation): $600M. Milestone (commercial launch): $400M. Profit split: 50:50. Development expenses and commercial costs: 50:50. Geographies: Teva (Europe, Israel), Sanofi (US, Japan, and RoW)
Is this a good deal for Teva?:
On the surface, the financials may pale in comparison to Merck’s $11B Prometheus buy-out. However, Prometheus had phase-II data in-house which Teva doesn’t, already providing a discount.
Sanofi is arguably the biggest immunology player and this may make a large difference from a commercial perspective. Analysts from Evercore believe Teva’s drug could be a $2B as base case with a regular pharma (given it may be third to market) or $5B base case with a major immunology player such as Sanofi backing it. So perhaps Teva could have received a larger upfront from a smaller player but they decided to make a bet on the commercial difference.
Other highlighted deals:
Automata $40M closer to building the automated lab of the future
SomaLogic and Standard BioTools merge to create a $1B multi-omics company
Regeneron and Intellia’s expanded CRISPR mission for neurological and muscular diseases
In case you missed it
AAV-based in vivo gene therapy for neurological disorders [Ling et al., Nature Reviews Drug Discovery, 2023]
What we liked on Twitter
Events
Biohack NYC [Oct 27th and Nov 11th, NYC]
Field Trip
Did we miss anything? Would you like to contribute to Decoding Bio by writing a guest post? Drop us a note here or chat with us on Twitter: @ameekapadia @ketanyerneni @morgancheatham @pablolubroth @patricksmalone








really like the deals/finance section. if you maybe could start a section, explaining finance stuff specifically unique to biotech, that would be super neat. thanks for your work!