BioByte 44: docking with AlphaFold, EvolutionaryScale, traversing chemical space, future prospects of LNPs, engineered antitumor bacteria, glial replacement in neurodegenration
Welcome to Decoding Bio, a writing collective focused on the latest scientific advancements, news, and people building at the intersection of tech x bio. If you’d like to connect or collaborate, please shoot us a note here or chat with us on Twitter: @ameekapadia @ketanyerneni @morgancheatham @pablolubroth @patricksmalone. Happy decoding!
Happy Friday! Here’s what we have in store for you this week:
More evidence around the poor accuracy of AF2-predicted structures in ligand docking experiments
EvolutionaryScale raises $40M
Decision making frameworks in biotech
COATI: a one-step generative method that can simultaneously explore chemical space while optimizing multiple properties of interest.
A review the efforts in motion to overcome the challenges and the clinical prospects of mRNA-LNPs
Young glial progenitor cells competitively replace aged and diseased human glia in the adult chimeric mouse brain
Engineered skin bacteria induce antitumor T cell responses against melanoma
What we read
Blogs
Docking With AlphaFold Structures: Oops [Derek Lowe, Science, August 2023]
Derek Lowe reviews a new paper studying the accuracy of AlphaFold binding mode predictions between small molecules and proteins. Recent protein structure-prediction models have shown admirable accuracy at both predicting structure from amino acid sequence and ‘inventing’ new protein structures that don’t exist in nature. However, small molecule binding prediction involves a larger number of structural motifs and interaction mode combinations. Proteins are made up of twenty amino acids and are connected by amide bonds. There are, therefore, a relatively small number of interaction modes and structure motif combinations for a protein. Small molecules, on the other hand, have many different types of motifs and interactions that don’t come up in proteins (e.g. halogen bonds, pi-interactions…). Given there are many more types of situations a model would have to learn from and there is only a relatively sparse data set in existence, predicting small molecules is a problem that is not currently set up to be solved with AlphaFold models.
This paper shows exactly that. The authors used AF2 on GPCRs, with no experimentally determined structural data allowed in the training, to predict their structure. They then used the predicted AF2 structure to predict where a ligand would bind using widely-used docking software. AF2 however did not give any improvement to other widely used techniques (e.g. homology modeling) when there is no ligand-protein structure that is experimentally elucidated.
AF2 is not the optimal model for many aspects of drug discovery. We do think that purpose-specific models could make huge advances in the docking-prediction field if we generate enough correct data as input.
Ex-Meta Researchers Have Raised $40 Million From Lux Capital For An AI Biotech Startup [Kenrick Cai and Iain Martin, Forbes, August 2023]
The former Meta protein-folding team, creators of ESMFold, is launching a new company called EvolutionaryScale. The startup will build protein language models that will test the scaling hypothesis of LLMs in biology - to what degree increasing the size of a model and training set improves performance and enables general purpose algorithms. The hope is that by training progressively larger protein language models, new capabilities beyond protein structure prediction will be unlocked, such as sequence to function mapping, or predicting how proteins interact with other biological molecules. Of note, scaling will be very capital intensive: “EvolutionaryScale projects that it will spend $38 million in its first year, with $16 million going to computing power, per the pitch document. Costs multiply from there, up to $161 million in year two and $278 million in year three (with $100 million and $200 million spent on compute, respectively). But throughout the document, the company repeatedly emphasizes that it could take ten years for biology AI models to help design products and therapies.”
On Decision Making Frameworks [Mason Victors, 2023]
Mason Victors, a biotech veteran and now consultant and founder of Idealist Warrior Labs published his first blog post on a decision making framework he heavily used in his techbio career, though the principle applies to all company building—DDCS—debate, decide, communicate, support. Mason walks through decision making frameworks including DDCS with some tips on implementing. A great read for company builders, though we’ll summarize a few points below:
Smaller organizations generally default to decisions by consensus. Makes execution easier since everyone is bought in but doesn’t scale well
Good people with strongly differing opinions leads to failure to reach consensus though a desire to reach consensus. Read: decision doesn’t get made because not everyone agrees.
Not making a decision is making a decision
Setting the stage: there has to be one single person who is appointed the decision maker (alignment on this is a prerequisite). Doesn’t necessarily mean the person with highest authority.
Debate: all opinions are heard. Not everyone needs to weigh in on debate.
Decide: single decision maker makes the decision. Be explicit about action resulting from a decision as well as action that won’t happen because of decision
Communicate: written records, why behind the decision, decision language
Support: back the decision once it is made. No “I told you so”
Being an effective team > being right
Overuse of frameworks can be equally as destructive.
Academic papers
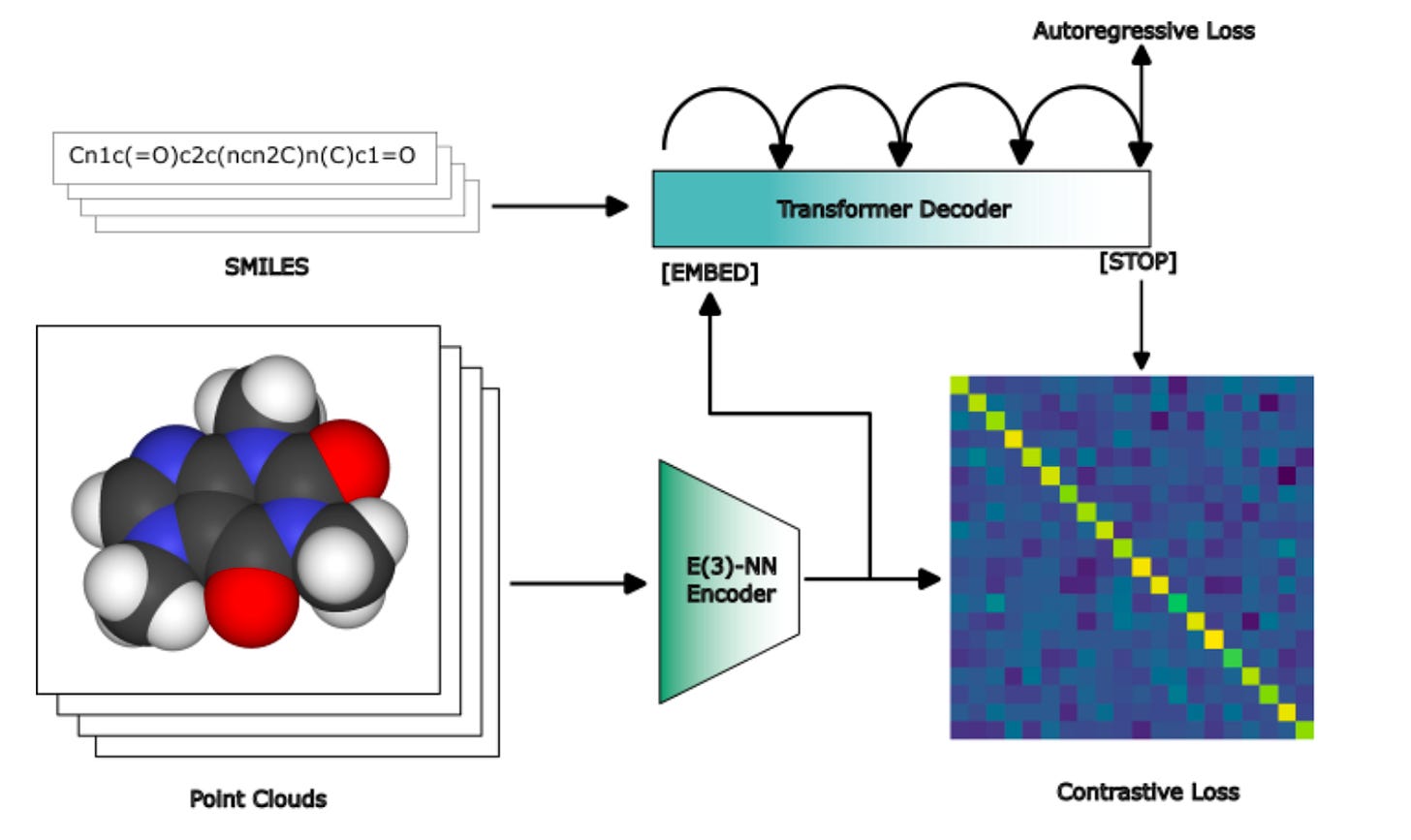
COATI: multi-modal contrastive pre-training for representing and traversing chemical space [Kaufman et al., chemRxiv, 2023]
Why it matters: Generative AI for small molecules is an incredibly tricky problem. It requires exploring a vast optimization space of over 10^60 drug-like small molecules, all while optimizing over multiple chemical properties. The traditional approach to generative AI for producing small molecule drugs typically follows two steps: 1) brute-force molecular generation using generative algorithms, followed by 2) molecular property prediction to filter for properties or functions of interest. While this approach has found some success, it is extremely computationally inefficient, and there’s no guarantee it will find the best molecule. This week, the team at Terray Therapeutics published a preprint detailing a better approach using a one-step generative method that can simultaneously explore chemical space while optimizing multiple properties of interest. The method (called COATI, or Contrastive Optimization for Accelerated Therapeutic Inference) comprises four integrated components:
High-quality, quantitative experimental data generated using Terray’s platform, which has generated 2 billion binding measurements
A molecular representation schema that is decodable (meaning any molecule can be explored directly in latent space and inverted to its chemical structure and synthesized for testing) and predictive of molecular properties
Differentiable regressors which permit exploration and optimization of chemistry in the representation space (think gradient descent to find local/global minima)
An algorithm for exploring and optimizing novel molecules in chemical space
The primary innovation of the paper was component #2: a unified, decodable representation space for small molecules generated via contrastive learning (a form of self-supervised learning in which unlabelled training samples are contrasted to learn which are similar/different) on multiple molecular modalities. A multi-modal encoder-decode model was trained on 2D text and 3D point representations of molecules using a contrastive pre-training task, but the framework is flexible enough to integrate additional data types.
The Terray team showed experimentally that COATI performs better than alternative methods like molecular fingerprinting. The method was also validated on a real-world application of multi-parameter molecular optimization of carbonic anhydrase. Going forward, one of the most exciting applications of COATI will be the ability to condition the generation of novel molecules on the basis of the COATI latent space with any desired property or function using experimental data, such as binding affinity. The only bottleneck - as always in drug discovery - becomes access to high-quality, relevant experimental data. For more details on how Terray is addressing this bottleneck, check out their profile in the Decoding Bio Report.
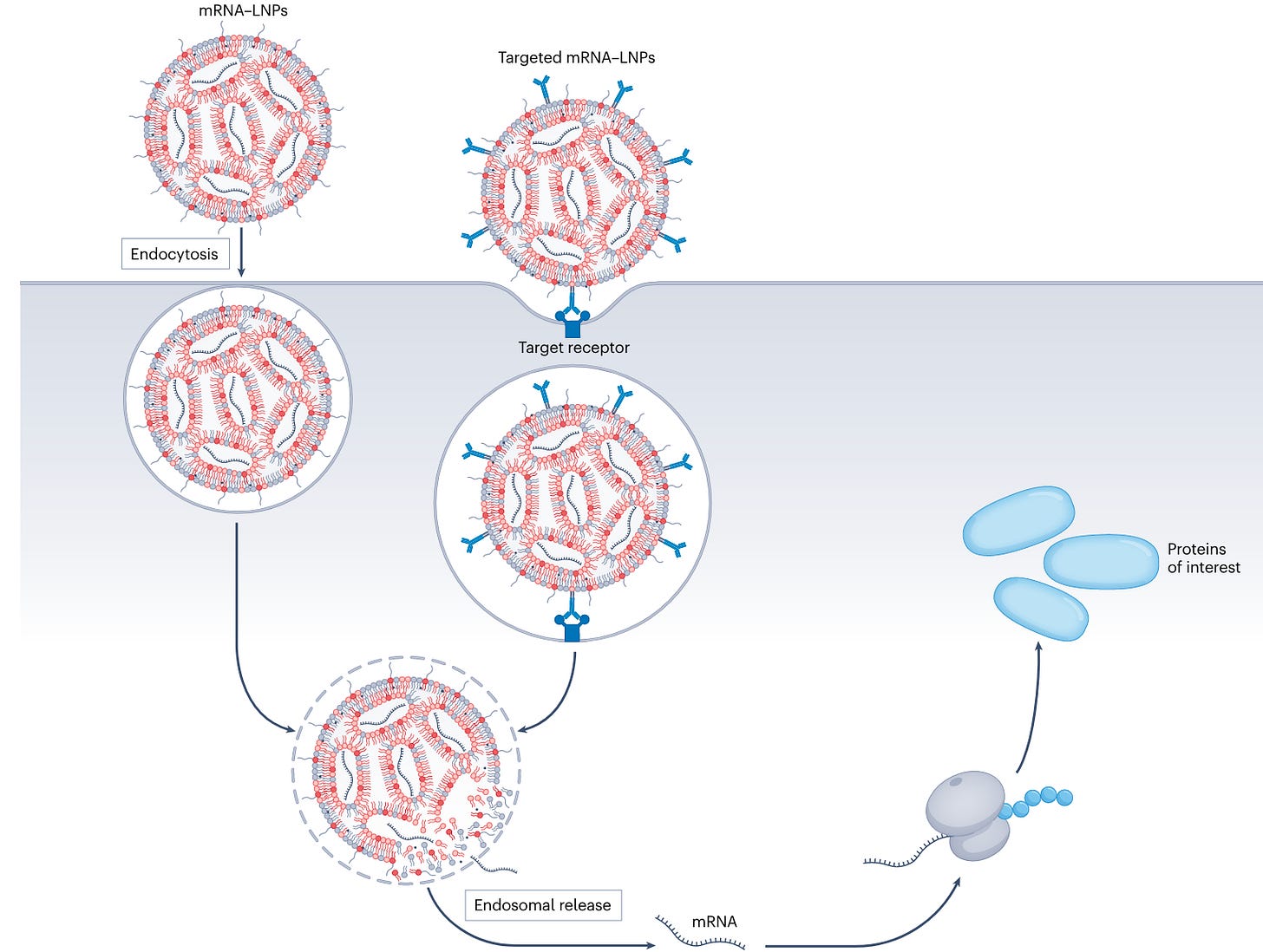
Targeting cancer with mRNA–lipid nanoparticles: key considerations and future prospects [Kon et al., Nature Reviews Clinical Oncology, 2023]
Why it matters: LNPs are a key component to the success of mRNA as modality in infectious diseases and a critical piece for its use in oncology. There are many challenges, however, ranging from manufacturability, targeting and immunogenicity. The authors review the efforts in motion to overcome the challenges and the clinical prospects of mRNA-LNPs.Biodistribution and transfection (TME permeability, efficiency of endocytosis and endosomal release, liver accumulation), immunogenicity and tropism depend on the formulation of the LNP, the nucleic acid payload and the method of administration. Given the huge search space these variables create, and the limited amount of publicly available data of in vitro and in vivo LNP assay readouts, there has been few successful efforts at predicting (and generating) LNP properties without using empirical methods. High-throughput experimentation includes in vitro and in vivo assays with various LNP formulations and tagged with DNA barcodes, which result in poorly correlated results when substituted with the ‘actual’ payloads.
However, significant advancements have been made over the last five years. There are several oncology mRNA-LNP therapies in the clinic, such as individualized cancer vaccines and intratumoral expression of immunoactivating cytokines. Therapies in the clinic apply passive targeting (that is, no specific additional components like mAbs on expressed on the lipid surface) which have a limited transfection efficiency when administered systemically and hence why they are used for vaccines where expression can be low for the therapy to be efficacious or administered intratumorally. Active targeting can increase the expression and can be used in systemic administration but come with cost and manufacturing challenges. The field is still young and requires systematic identification of LNPs that can deliver payloads at high efficiency, be tissue-specific and evade the immune system. Finally, the authors call to action to all mRNA-LNP developers to think about CMC early in the therapeutic development whilst deciding treatment design.
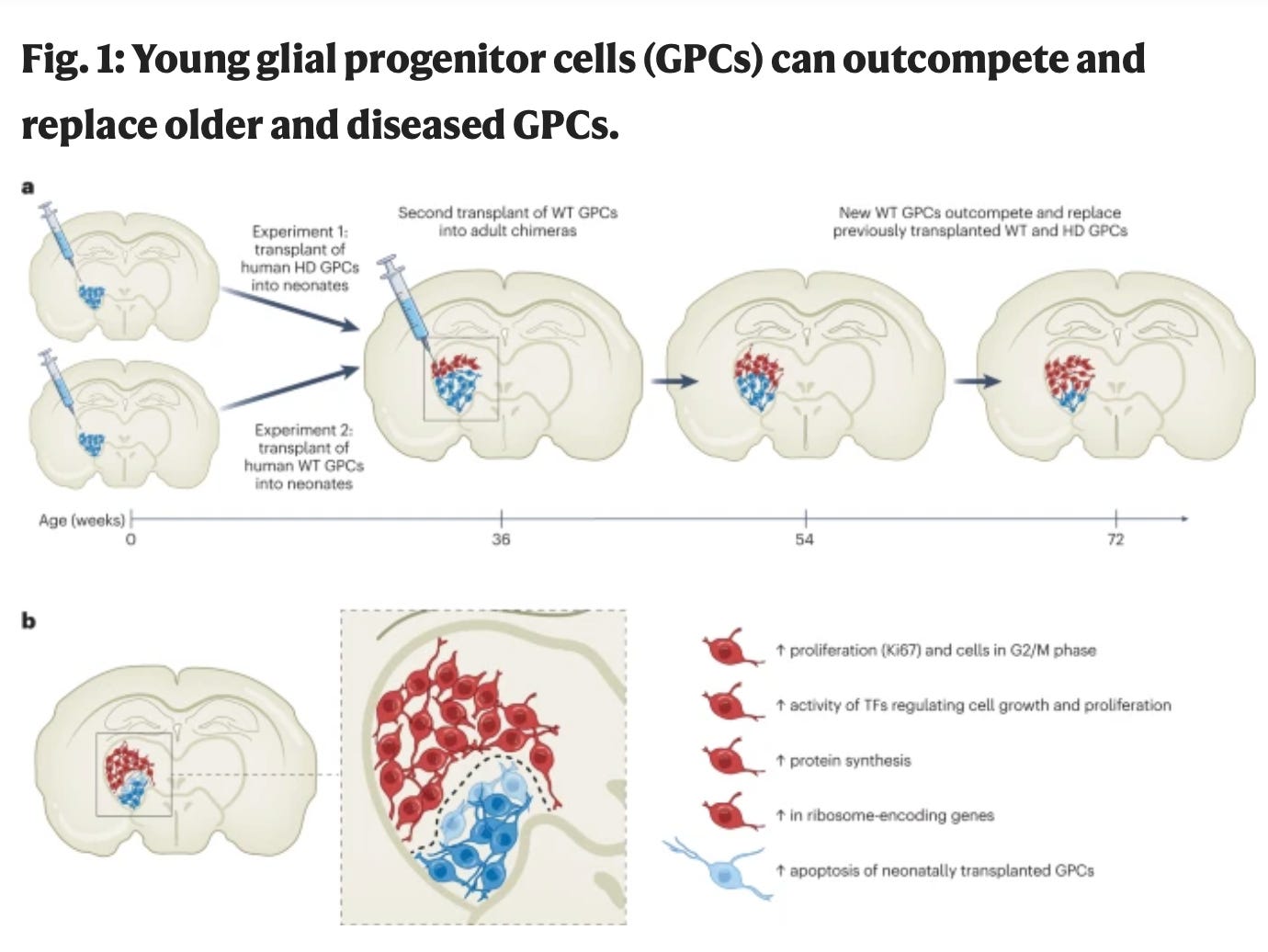
Young glial progenitor cells competitively replace aged and diseased human glia in the adult chimeric mouse brain [Vieira et al., Nature Biotechnology, 2023]
Why it matters: Glial cells, which serve as supporting cells in the central nervous system, have essential functions in upholding homeostasis within the CNS. Lately, more attention has been drawn to the involvement of abnormal glial cells in the advancement of neurodegenerative disorders such as Alzheimer's and Huntington's diseases, among others. In this paper, the authors demonstrate that transplantation of human glial progenitor cells (GPCS) outcompete and replace impaired human glial cells affected by Huntington's disease or the aging process. This research highlights the potential for a cell-based therapeutic approach to address brain-related neurodegeneration and aging.Deranged glial cells can result in neuronal damage via the loss of cell-cell support, pro-inflammatory cytokine productions, and disruption of the blood brain barrier. Glial GPCs are ubiquitous throughout the CNS and are responsible for spontaneous production and replacement of glial cells over the course of life. Here, the authors generated GPCs from Huntington’s embryonic stem cells and transplanted them into the striatum of mice. 8 months later, these cells took over almost the entire striatum, creating a chimeric mouse brain. They subsequently transplanted healthy GPCs into mice and found that healthy GPCs eliminated the majority of resident Huntington’s GPCs. To ascertain whether this was a function of disease state or cell age, the authors performed neonatal implants of healthy GPCs into mice, followed by a second transplant of healthy GPCs upon maturity. They found that the younger GPCs outcompeted healthy resident aged cells, suggesting that competitive advantage depended on the relative ages of competing populations, rather than the presence of disease phenotype.
The authors used single-cell RNA-seq and found that dominant younger cells had higher expression of genes involved in proliferation and protein synthesis, while the aged GPCs were more often undergoing apoptosis (which only increased upon challenge with young GPCs). This work is exciting as it suggests that aged and diseased human glia may be replaced with a transplant of younger GPCs, opening up the doors for a new therapeutic approach of challenging brain states.
Engineered skin bacteria induce antitumor T cell responses against melanoma [Chen et al., Science, 2023]
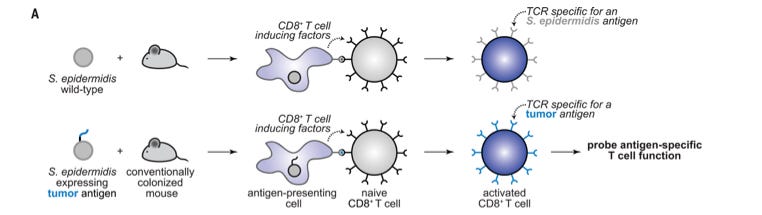
Why it matters: imagine fighting skin cancer with a lotion. The skin microbiome already provides a diverse array of bacteria that are commensal, or coexist, with the body. While most don’t trigger an immune response, this study focuses on engineering bacteria that can trigger a specific adaptive immune response to present tumor antigens and wire the bacteria to fight cancer accordingly. This research introduces a new therapeutic approach for treating melanoma. This group is not the first to think of using bacteria as cancer therapeutics. The researchers took the skin bacterium Staphylococcus epidermidis which naturally gives rise to an adaptive immune response and engineered them to express tumor antigens anchored to cell-surface proteins. They engineered bacteria with E. coli based plasmids and used both heat shock and electroporation. As a control, ovalbumin was used as a model antigen because the antigenic-presentation is well-characterized. The engineered bacteria secreted T cells that could then circulate lesions for anti-cancer effect, both locally and to metastatic sites. This last finding is particularly interesting because it shows that the bacteria not only stimulate relevant antigen specific T cells that slow the progression of melanoma, but they can do so at distal tumor sites, which for melanoma is important due to the quick growth of cells. Follow up experiments show the bacteria’s synergy with the immune checkpoint blockade, contributing to immune memory for the antigens.
What we listened to
#51 – Robert Sapolsky, Ph.D.: The pervasive effect of stress – is it killing you?
Notable Deals
Startup Cellares adds $255M as investors pour cash into cell therapy production
J&J-backed startup raises another $150M for brain drug development
Royalty Pharma pays Ferring $300M for 5% royalty on sales of cancer gene therapy Adstiladrin
In case you missed it
Nvidia exec: Generative AI can turn every biologist into a computer scientist — and vice versa [Brian Buntz, Drug Discovery Trends, August 2023]
What we liked on Twitter
Events
London’s Generative AI x Healthcare Hackathon
Field Trip
Did we miss anything? Would you like to contribute to Decoding Bio by writing a guest post? Drop us a note here or chat with us on Twitter: @ameekapadia @ketanyerneni @morgancheatham @pablolubroth @patricksmalone










Loved the decision making frameworks! Thanks for sharing 🧬