BioByte 48: review of the obesity drugs, wearable non-invasive sensor for measuring female hormones, building a virtual cell, cracking the chemical code, is target-based drug discovery a waste of time
Welcome to Decoding Bio, a writing collective focused on the latest scientific advancements, news, and people building at the intersection of tech x bio. If you’d like to connect or collaborate, please shoot us a note here or chat with us on Twitter: @ameekapadia @ketanyerneni @morgancheatham @pablolubroth @patricksmalone. Happy decoding!
Happy Autumn! Thought it doesn’t feel like it, we’re officially one year into BioByte. To be totally honest, our goal was to make it to BioByte12 so putting out 48 feels really good. We’re also brainstorming what we can do with Decoding Bio outside of BioByte, so if you have a wishlist, drop us a note.
It’s a long one today. Here are the highlights so you can pick your own adventure:
when the obesity drugs work too well…
is target based drug discovery a waste of time
reviewing the CZI plans to build a virtual cell
when the genetic code falls short…
protein models x evolution
deep brain stimulation for depression recovery
continuous monitoring of female hormones via noninvasive nanotechnology
What we read
Blogs
When the Breakthrough Obesity G-Agonist Drugs Exceed Expectations [Eric Topol, Ground Truths, 2023]
G-agonists, which mimic endogenous G-protein-coupled receptor peptides called incretins, are on track to become the most successful drugs in medical history, even more than life changing statins.
Incretins are gut-secreted hormones that include GLP-1, GIP and GCG, which have overlapping but also distinct effects on the gut and brain with respect to metabolism, inflammation and appetite.
What’s new?
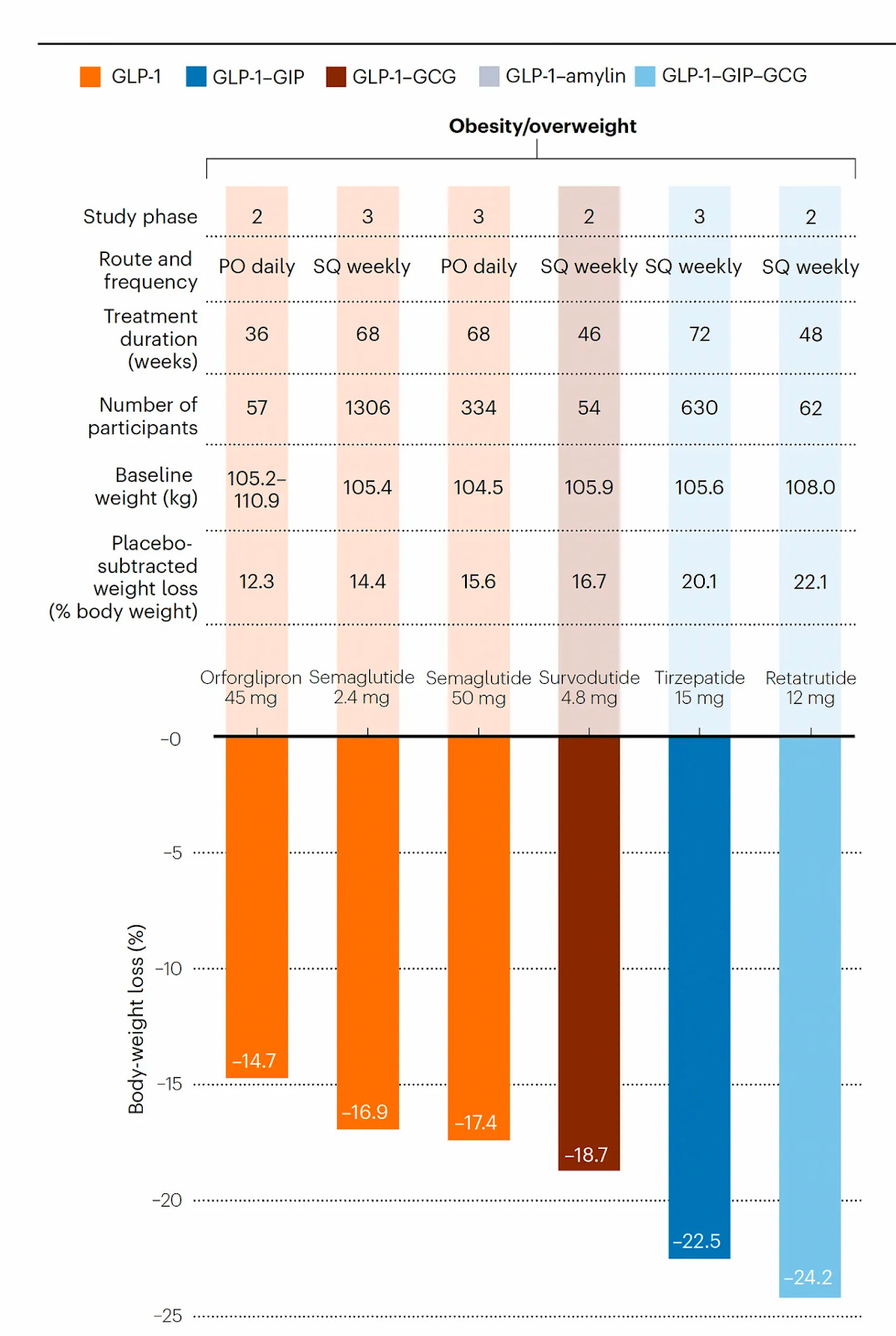
The first triple incretin inhibitor (retatrutide) recently completed its Phase II trial (with results on weight loss shown below). The efficacy speaks for itself. Importantly however, at the highest dose 16% of participants had to discontinue the drug due to GI side effects and nearly 40% of the weight loss was due to loss of lean body mass.
Further evidence shows that semaglutide can decrease major cardiovascular events (by 20% according to Novo’s trial), but the data is yet to be published.
A 1 year follow up study showed that a measurement of blood glucose (HbA1c) and C-Peptide improved considerably in Diabetes Type 1 patients when using semaglutide. Even if a small minority of Type 1 diabetics discontinued insulin use, it could be a huge life changing moment.
Despite the success and efficacy of the drugs, we don’t know exactly how these drugs work. For example, why do people with diabetes lose less weight than obese, non-diabetics? Despite the target-based discovery, there seems to be an unexplainable difference when looking at the clinical phenotypes. This leads in nicely into Lowe’s summary below…
Target Based Drug Discovery - A Waste of Time? [Derek Lowe, Science, 2023]
A new paper authored by Arash Sadri claimed that only 11% of approved small molecule drugs were discovered by purely target-based assay methods, and the rest have been discovered through phenotypic means. Sadri took on the odyssean task of reviewing the origins of all 1144 small molecule drugs, by interviewing people working in those projects amongst other means.
So what is going on?
As Lowe mentions: “Every living creature is the end result of billions of years of phenotypic optimization”
Even for drugs with were discovered via target-based approaches, their clinical utility was enhanced by unanticipated effects observed phenotypically after the fact
Survivorship bias at work: many drugs fail at Phase II trials, and the ones that get through are not the ones with most selectivity or potency for their stated targets. Most drugs are polypharmacological in mechanism, regardless if the drug was found via a target-based approach.
The apparent recent increases in productivity have been due to the adaptation of the industry to its failures rather than addressing their fundamental causes. I.e. targeting monogenic diseases with new modalities
Chan Zuckerberg Science To Build AI GPU Cluster To Model Cell Systems [Chan Zuckerberg Initiative, September 2023]
The Chan Zuckerberg initiative recently announced plans to create a "virtual cell" to support biomedical research. An AI-based simulation of healthy and diseased cells will be developed using a high-performance computing cluster of 1000+ GPUs, some of the most powerful compute infrastructure ever built for non-profit research. While exciting, it is important to learn from past mistakes of similar initiatives to ensure this project is maximally successful.
Consider the Human Brain Project, a decade-long, $600M+ project with the goal of developing a simulation of the human brain. While the initiative produced some important publications and resources, it largely failed to deliver on the overarching goal of combining large datasets and computational methods to simulate the function of the whole brain. In many ways, the project was destined to fall short because simulating a brain is not a realistic objective given the state of contemporary neuroscience. Worse, there was little alignment amongst the hundreds of scientists involved on how best to make progress towards this goal, and the project changed leadership and strategic direction many times. With the initiative wrapping up this month after a 10 year journey, the project has produced fragmented tools and resources rather than a unified theory or simulation of brain function.
What can initiatives like Chan Zuckerberg's learn from the Human Brain Project? A few ideas:
The lofty goal of simulating a cell should be balanced with smaller-scale, focused research projects that are more tractable. It is reassuring to see specific scientific goals mentioned for the CZI initiative, such as developing models for predicting cell types and states.
Clear leadership and a transparent, unified vision for the project's strategy and goals
Efficient resource utilization, especially compute and funding, is critical. The risk of mega-science projects like this one is the opportunity cost of funneling resources into one large project vs many smaller ones.
The Chemical Code: An Opportunity Bigger than the Genetic Code [Viswa Colluru, Enveda Biosciences, September 2023]
Viswa takes to the Enveda blog to pen his thoughts on decoding biology one step further than genetics. He brings up the excellent point that we’ve been so floored by genetic advances (which truly are amazing) that we’ve forgotten our understanding of the genetic code is not necessarily 100% complete and it is not the only way to view biology. In fact, zeroing in on this alone renders our picture of life itself incomplete. Instead, Viswa argues we should be looking at the chemical code making the point that genes, RNA, proteins all end up coding for things that do work—transferring energy and matter. Thus, chemistry is an underlying force stringing life together and our current understanding of the chemical code is severely lacking. Enveda is doing really cool work trying to crack this code by focusing on metabolomics. We won’t dive into it here but highly recommend reading Viswa’s full post to get a sense for how much we have left to discover and how one company is tackling the problem.
“Information isn’t life…a dead cell and a live cell have the same genome”. Instead, he contends, that metabolic flux —- the sum of continuous transformations of small molecules on a timescale of nanoseconds, nanosecond after nanosecond —- is the definition of being alive.
Academic papers
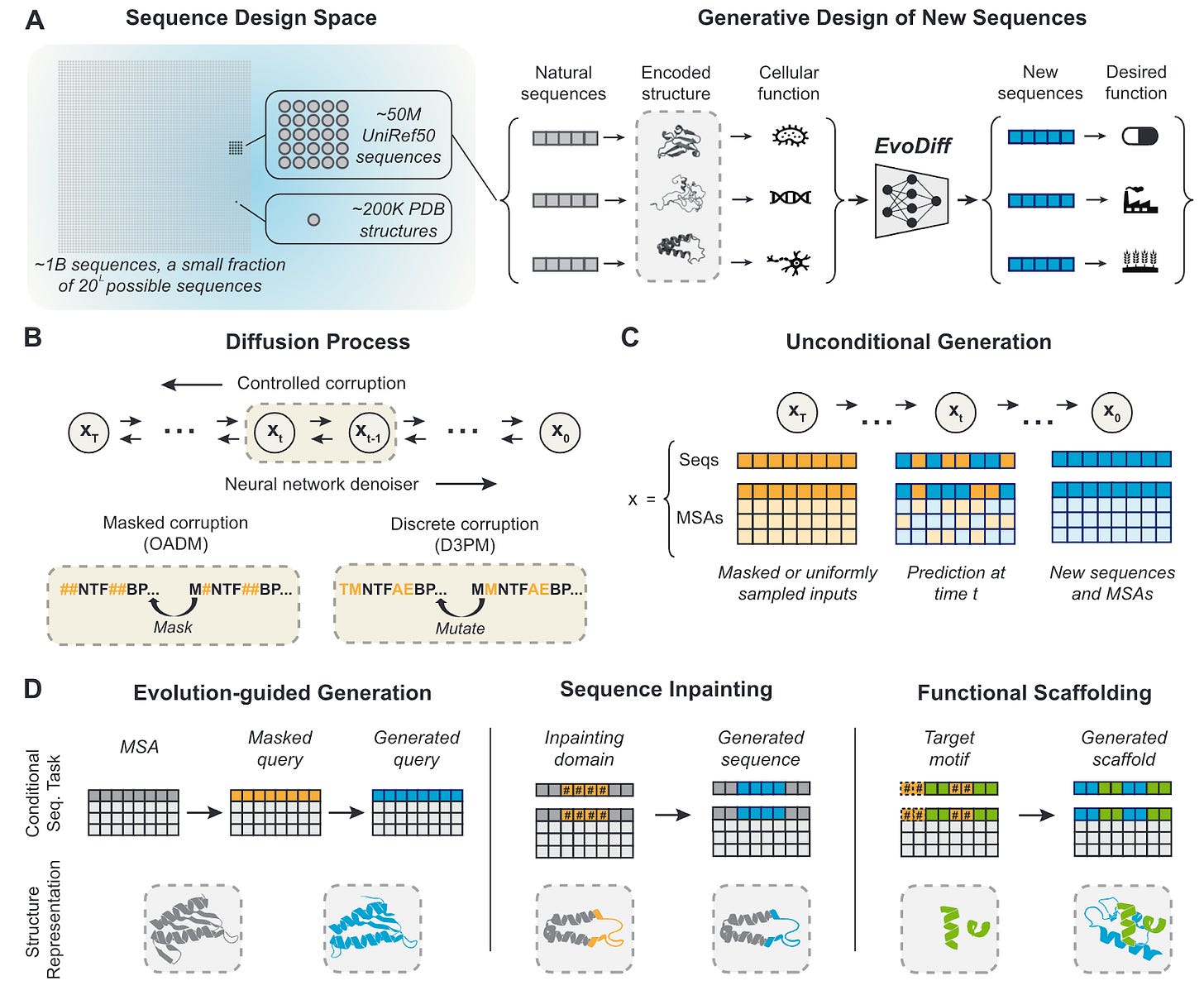
Protein generation with evolutionary diffusion: sequence is all you need [Alamdari at al., bioRxiv, 2023]
Why it matters: Evolution has only sampled a tiny fraction of protein sequences, and there are even fewer experimentally-determined structures. The authors have developed a new model trained on natural protein sequences that can output diverse, novel and realistic protein sequences. Diffusion models in protein design have been successful in outputting protein structures, rather than sequences.The authors present EvoDiff, a discrete diffusion model that is trained on protein sequence alone rather than their contemporaries which utilize structure data. Diffusion models which output structures such as RFdiffusion, limit the scope of the training data and restrict generation to small protein design space. EvoDiff can generate proteins inaccessible to structure-based models such as proteins with disordered regions.
EvoDiff uses a discrete diffusion framework in which a forward process iteratively corrupts a protein sequence by changing its amino acid identities, and a learned reverse process predicts the changes made at each iteration.
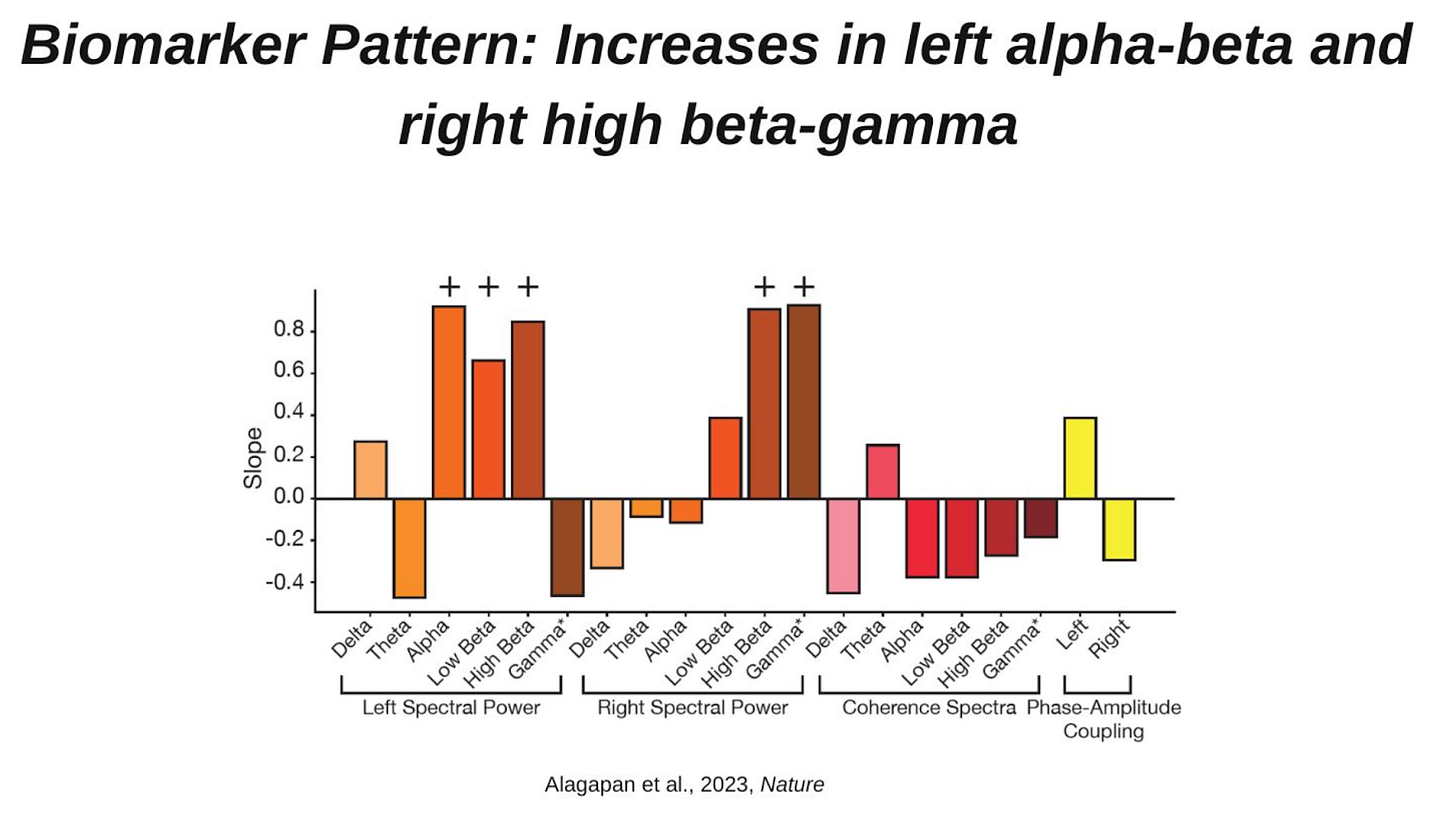
Cingulate dynamics track depression recovery with deep brain stimulation [Alagapan et al., Science, September 2023]
Why it matters: Deep brain stimulation for depression has previously failed in clinical trials due to a lack of precision stimulation customized to each patient. A new paper from a pioneer of DBS discovers neurophysiological signals that guide successful treatment, which can be incorporated into closed-loop DBS systems. Deep brain stimulation (DBS) for depression has had a roller-coaster history. A big part of the variation of therapeutic effect is due to the error-prone and unsystematic nature of programming stimulation parameters of the DBS device for each patient. In a new paper in Nature, DBS pioneer Helen Mayberg and collaborators at Emory and Georgia Tech published a study of 10 patients with treatment-resistant depression implanted in the subcingulate cortex (an area of the brain that helps regulate emotional behavior) with Medtronic’s closed-loop Activa DBS system. 90% of participants demonstrated robust clinical response, and 70% achieved remission. Using local-field potentials (an electrophysiological measure of the aggregate synaptic activity for a local population of neurons), a biomarker that predicted patient response was discovered that was present in all patients that responded to treatment. Interestingly, there wasn’t one straightforward electrophysiological signal that predicted treatment response, but rather a combination of signals across frequency bands (figure below; most predominantly in left alpha-beta and right high beta-gamma). The results need to be replicated in a larger sample, but these are promising objective biomarkers to help guide DBS of depression.
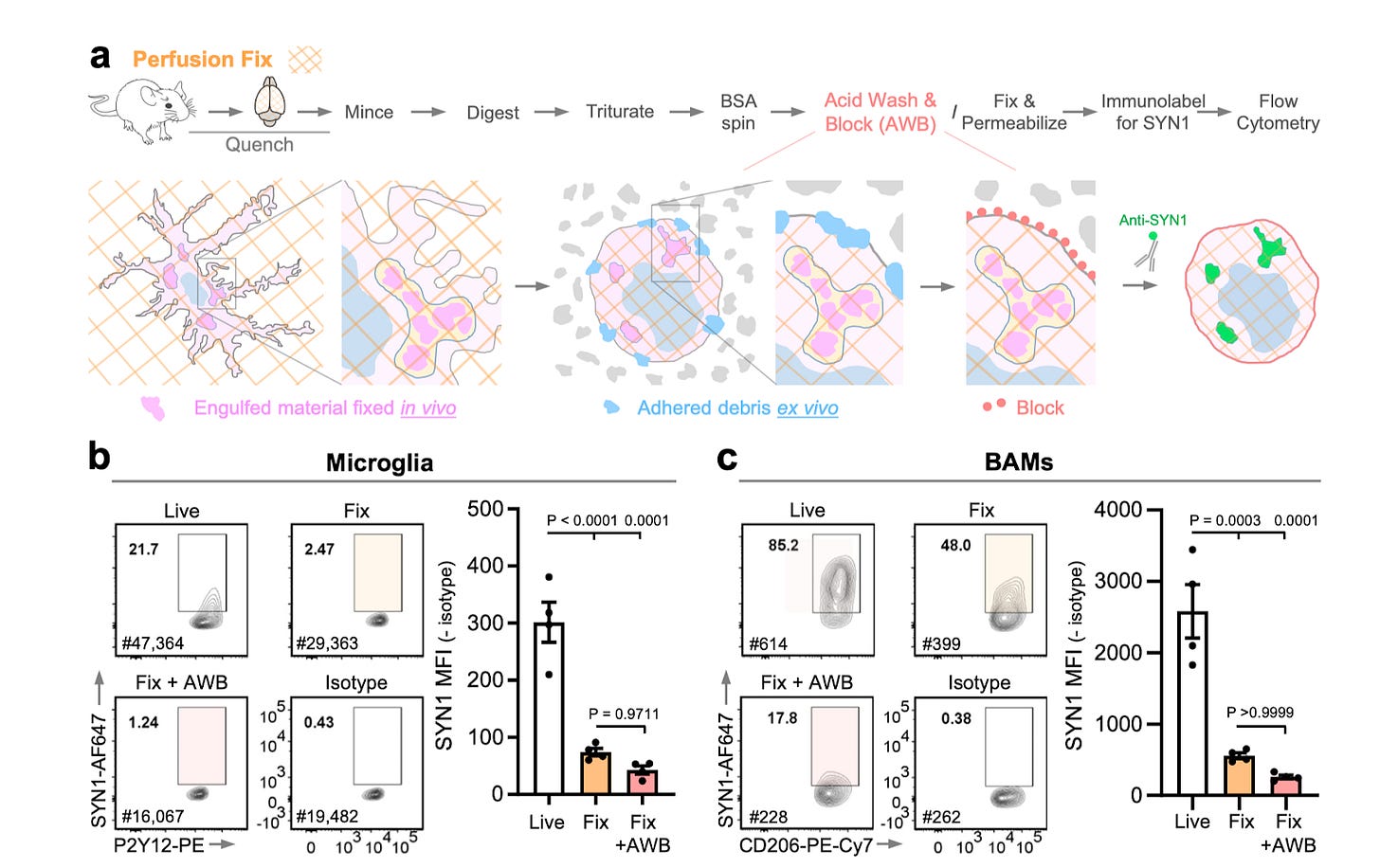
FEAST: A flow cytometry-based toolkit for interrogating microglial engulfment of synaptic and myelin proteins [Dissing-Olesen et al., Nature Communications, 2023]
Why it matters: Microglia play critical roles in CNS development and homeostasis, and their engulfment of synaptic material plays a critical role across health and disease. To date, there has been little success in high-throughput quantification of the engulfment process, which has limited our understanding of such mechanisms. Here, the authors develop FEAST (Flow cytometric engulfment assay for specific target proteins), which enables high-throughput, single-cell interrogation of in vivo engulfment of synaptic material via brain resident macrophages. Briefly, microglia are allowed to engulf a pre-labeled material (via fluorescent label, antibody-tagged, or genetically encoded fluorescent protein). Of note, it was found that fluorescent proteins needed a pKa lower or roughly equivalent to that of the lysosomal pH for stability. Immunohistochemistry demonstrated that these are retained within microglia, which is necessary for downstream flow cytometry. The microglia are are then isolated: for live microglia, cold Dounce homogenization (mechanical disruption) was used. However, to quantify endogenous synaptic and myelin proteins, the microglia needed to undergo enzymatic digestion and fixation/permeabilization in combination with a number of engulfment and lysosomal inhibitors to reduce ex vivo acquisition of debris and degradation of material. There was significant false positive signal; the FEAST protocol was subsequently modified to allow isolation of microglia from perfusion-fixed brain itself (fixing in vivo eliminates concerns of ex-vivo engulfment and allows removal of contaminating debris with an acid wash before labeling engulfed material). This significantly improved the ability to sensitively and specifically detect differences in microglia engulfment of synaptic proteins across three experimental models. FEAST can be readily expanded to quantify engulfment of any material by any cell type; this technique can be used to glean critical information about cellular dynamics that shape homeostasis and disease.
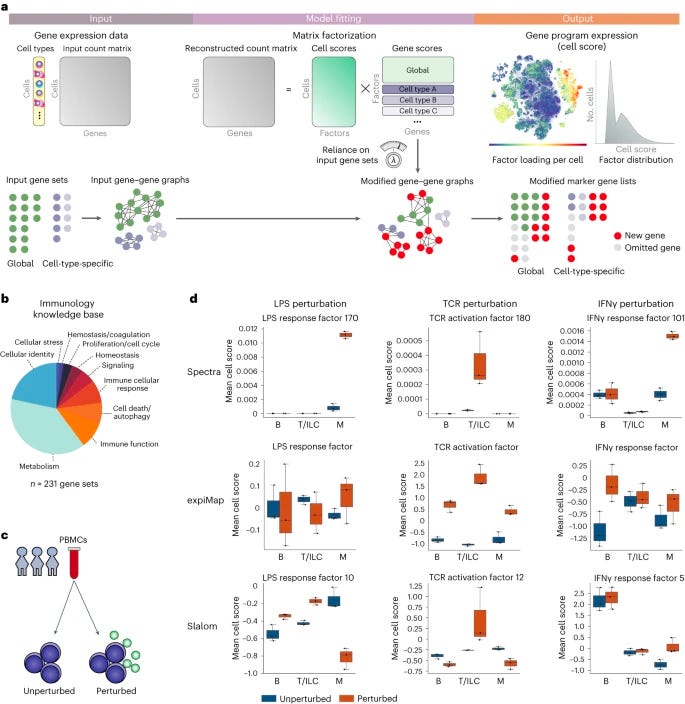
Supervised discovery of interpretable gene programs from single-cell data [Kunes et al., Nature Biotechnology, September 2023]
Why it matters: Interpreting single-cell data to find meaningful gene programs remains challenging. Spectra provides a powerful approach to leverage prior knowledge while allowing data-driven discovery. The ability to separate highly correlated biological processes like T cell exhaustion and tumor reactivity enables new insights into cell state and function. Reproducible discovery of gene programs opens up many downstream applications for translational insights and biomarker identification. Spectra strikes a balance between incorporating prior knowledge and unbiased learning that could enable broad adoption for analyzing single-cell data across diseases.The paper presents Spectra, an algorithm for analyzing single-cell RNA sequencing data to identify interpretable gene programs. Spectra uses prior biological knowledge like gene sets and cell types to guide discovery of factors that represent cellular processes. It represents this knowledge as a gene-gene graph. The algorithm decomposes expression into a minimal set of factors that reconstruct the data and match the input graph. It can modify the graph and find entirely new factors. Spectra outperforms previous methods at recovering simulated ground truth factors and real gene sets. It scales efficiently to large datasets.
Analysis of tumor immunology data shows Spectra can separate highly correlated T cell exhaustion and tumor reactivity programs. It identifies a tumor reactivity factor associated with immunotherapy response. Spectra reveals metabolic pathways like lysine metabolism in specific immune cell types. It finds a pro-metastatic macrophage program that expands in immunotherapy non-responders. Factors are reproducible across independent breast cancer datasets and a large lung cancer cohort, associating with clinical variables. This demonstrates robustness to batch effects.
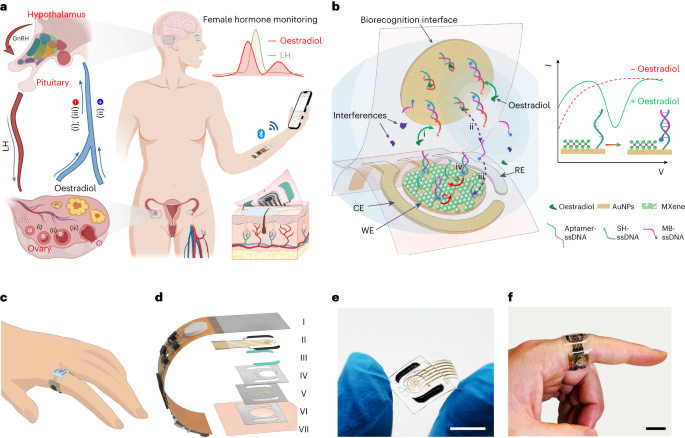
A wearable aptamer nanobiosensor for non-invasive female hormone monitoring [Ye et al., Nature Nanotechnology, September 2023]
Why it matters: continuous monitoring of most hormones is not yet possible. Given that hormones are constantly fluctuating and these fluctuations have a drastic impact on health and lifestyle, a noninvasive way to check levels can have implications ranging from fertility to family planning to diagnostics. This tech is a breakthrough in the ability to use aptamer technology at a high enough resolution to read female hormone fluctuations. This team from Caltech designed a skin-interfaced wearable aptamer nanobiosensor that automatically monitors estradiol through sweat analysis. It uses a gold nanoparticle electrode to get high sensitivity at low levels of detection, where other groups have perviously struggled. Oestradiol measurement requires pM-scale detection and this group’s sensor goes to a detection limit of 0.14pM. The researchers validated the device in human participants. When checked against serum concentrations of estradiol, similar trends and results were seems in the sweat concentrations measured by the device. This data was collected over the course of the menstrual cycle and further supports the use of sweat as a tool for measurement. The device itself is presented as a bendable wrap that sits on your finger, similar to a ring. It’s unclear how the form-factor may change with future iterations or whether other sensors can be integrated into the device for more comprehensive measurement.
What we listened to
Notable Deals
Jura Bio launches with $16M for AI x cell therapy
LB Pharmaceuticals raises $75M for schizophrenia drug as Karuna looks to change the field
Seraxis closes $50M for diabetes program
Samsung Biologics engages in $242M BMS deal
Valo engages with Novo Nordisk for cardio metabolic programs with upfront of $60M
AIRNA, an RNA-editing startup, launches with $30M
In case you missed it
Endpoints 11: Biotech’s most exciting startups
Chan-Zuckerberg AI Cell initiative
Is AlphaFold really the next big thing
Events
BioML Seminar with Peyton Greenside of Big Hat Biosciences in Berkeley, CA
Field Trip
Ringing in autumn and staying relevant with this classic…
Did we miss anything? Would you like to contribute to Decoding Bio by writing a guest post? Drop us a note here or chat with us on Twitter: @ameekapadia @ketanyerneni @morgancheatham @pablolubroth @patricksmalone