BioByte 53: A new AlphaFold model, Future House to accelerate science, the shallow brain hypothesis, direct conversion of carbon to nitrogen, Lilly likes cardiovascular disease
Welcome to Decoding Bio, a writing collective focused on the latest scientific advancements, news, and people building at the intersection of tech x bio. If you’d like to connect or collaborate, please shoot us a note here or chat with us on Twitter: @ameekapadia @ketanyerneni @morgancheatham @pablolubroth @patricksmalone @zahrakhwaja. Happy decoding!
Winter is here. Are you on a gondola in Jackson Hole about to rip some slopes? Mid-sleighing with the dogs in Minnesota? Is it 1AM and you’re waiting for the headlining act to show up at 3 AM at the Brooklyn Mirage? Well lucky for you, we got some science:
What we read
Blogs
A new AlphaFold model: in AI for biology, you get what you train for.
Since the release of AlphaFold in 2020, a once-hypothesized revolution in drug discovery has failed to materialize. While accurate at predicting protein structure, the utility of AF for drug discovery has been limited. New AF models published this week by Isomorphic Labs (see overview in the Paper section below) change this.
A useful case study for understanding the difficulty of translating protein structure algorithms like AF to drug development is molecular docking. Docking is a computational method for predicting the orientation and position of a ligand when it binds to a protein with the goal of generating new small molecule drugs, and is the workhorse of structure-based drug design. The docking process requires the crystal structure of a protein bound to ligands, however these structures are expensive/difficult to generate.
There was a lot of excitement after AF’s release about replacing crystal structures with AF in docking in order to discover drugs more quickly and cheaply. Many expected this to work because AF-generated structures closely match the crystal structures of protein binding pockets. However, several recent papers (Karelina et al., 2023, He et al., 2022, Heo and Feig, 2022) have found that despite AF’s ability to predict protein structure, the accuracy of molecular docking decreases significantly when using AF-generated structures.
Why didn't AF-generated structures translate to an ability to predict drug binding? Generating a structural model for a small molecule binding to a protein turns out to be a much more difficult problem than protein structure alone. The space of possible small molecules (10^60) and structures is much larger than that of proteins. Further, ligand binding changes protein structure, and AF wasn't trained on ligand-protein complexes.
A white paper out this week describes a new iteration of AF that achieves stronger performance on molecular docking. The paper is intentionally short on methodological details, but from what we can tell, new AF models were trained on protein complexes with non-protein elements, including small molecule ligands. The model achieves state-of-the-art accuracy on molecular docking benchmarks, outperforming both non-ML and ML based methods. importantly, comparison methods used ground truth bound protein crystal structures as input.
What's the takeaway? One important, obvious lesson is that in AI for biology, you get what you train for. To predict ligand-protein interactions, you must train on ligand-protein complexes. To predict antibody-antigen interactions, you must train on antibody-antigen complexes. Simply training on proteins (first generation of AF) is not sufficient for many applications in drug discovery, where the goal is to model complex interactions between many biological molecules. The capabilities of AI models in biology will only continue to advance as we incorporate specific and relevant biological data into model training to bridge the gap towards real-world impact.
Announcing Future House [Sam Rodriques]
Future House is a new non-profit focused on building AI systems to accelerate scientific discovery. Their 10-year mission is to create semi-autonomous AI Scientists to help human scientists be more productive. They are focusing on biology because it has the most potential to benefit humanity through advances in medicine, food, and climate science. Biology research is bottlenecked by the limited time individual scientists have.
Future House believes the components for an AI Scientist exist today thanks to advances like language models. However, human scientists will be critical to train and validate the AI. They will have biologists working closely with AI researchers.
Measuring progress will involve testing the AI on tasks in a "Challenge Book" from simple to complex scientific reasoning. Future House will pioneer a new approach as an independent non-profit to maintain focus on the long-term mission. They will have integrated, cross-disciplinary teams of biologists and AI researchers. Backed by Eric Schmidt, Future House also has an exceptional team including heads of science, engineering, and operations.
Second recipient of genetically modified pig heart dies, six weeks after surgery [Deborah Balthazar, STAT News, Oct 2023]
A 58-year old man with terminal heart disease underwent an eight hour surgery to receive a genetically modified pig heart—only the second human to undergo such an experimental xenotransplantation. He was well after surgery but began to show signs of organ rejection a few weeks later and passed away six weeks after the surgery at the University of Maryland. You may recall a similar story from last year, when the same team performed the same xenotransplantation procedure on another 57 year old patient who died two months after the surgery, though that cause of death may have been from an infection from a pig virus. Though far from a meaningful sample size, both organ failures cast some doubt into xenotransplantation, notably chances of organ rejection, viral infection, and other associated causes for failure. This surfaces interesting discussion on the ethics of continuing such experimentation knowing the last two trials failed but also knowing the patients have end-stage heart disease and were otherwise ineligible for transplant.
Academic papers
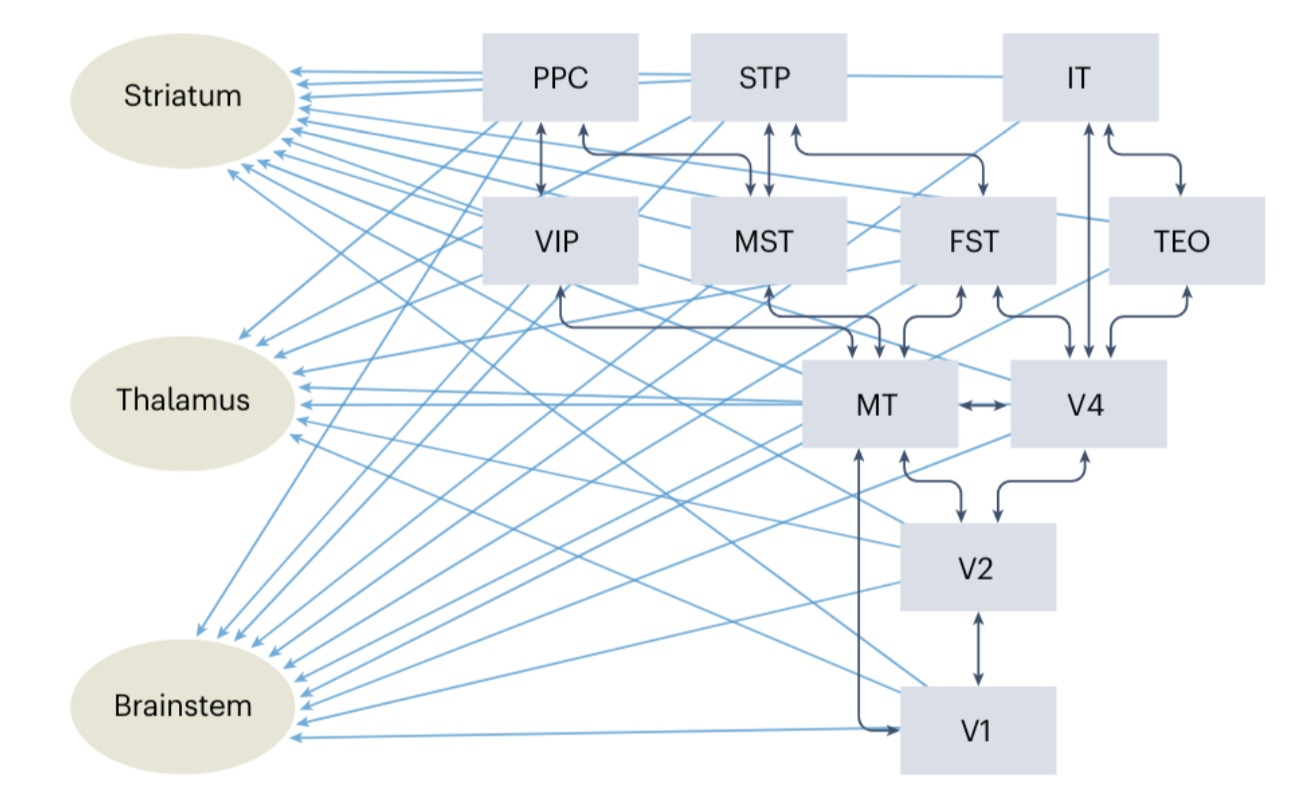
How deep is the brain? The shallow brain hypothesis [Suzuki et al., Nature Reviews Neuroscience, October 2023]
Why it matters: The authors of this paper posit that deep learning and predictive coding (e.g. Bayesian inference) architectures ignore neurobiological evidence that all cortical areas project to and receive signals directly from subcortical areas. They state that dominance of hierarchical architecture in deep learning is questionable, missing critical computational principles the brain uses.They present what the authors call “the shallow brain hypothesis”, where “hierarchical cortical processing is integrated with a massively parallel process to which subcortical areas contribute”. One of the key views that has led to this predominant view has been the notion that deep architectures perform better than shallow ones, or that they are required to solve certain problems (such as non linearly separable classification problems. However, two-layer restricted Boltzmann machines which use a shallow architecture, have demonstrated that they can perform to the same standard benchmarks.
Caption: Hierarchical, deep architecture
Caption: Shallow architecture
In the shallow brain hypothesis, the entire cortex is one giant layer that provides contextualized input to subcortical motor and premotor centers in the brainstem and spinal cord. Each of the cortical areas is reciprocally, and parallelly connected with subcortical areas, creating a shallow layer, rather than information passing layer to layer until there is one final output that connects with subcortical areas. There are some potential advantages to shallow networks: more efficient reinforcement learning and faster information processing.
Performance and structural coverage of the latest, in-development AlphaFold model [Deepmind, Isomorphic Labs, 2023]
Why it matters: generalizable models that can extend beyond protein structure prediction can be useful in the early stages of drug discovery to predict where small molecules and other ligands may bind. Docking models require reference protein structures, so being able to accurately model small molecule binding for proteins that do not have a bound protein structure available can be a practical starting point to understand how to drug a particular target.The teams at Google Deepmind and Isomorphic Labs have published a results-focused update describing the latest developments in AlphaFold. The new version of AlphaFold named AlphaFold-latest in this update, greatly expands on the range of applications of the model to carry out joint structure prediction of complexes including small molecules, proteins, nucleic acids, ions and post-translational modifications. It can also generate predictions for nearly all proteins in PDF, many with atomic accuracy.
It also shows increased accuracy over previous specialist models in four categories:
Without the use of ground truth bound protein structures, AlphaFold-latest outperformed the state of the art specialist tools for ligand docking.
Protein-protein structure prediction is improved versus AlphaFold2.3, especially in antibody-protein.
Protein-nucleic acid interaction prediction is also improved over RoseTTAFold2NA; yet still slightly worse at RNA structure prediction over the top CASP15 entrant
It is able to predict the structure of post-translational modifications and bounded ligands.
Carbon-to-nitrogen single atom transmutation of azaarenes [Woo et al., Nature, 2023]
Why it matters: Single atom changes can alter molecular properties significantly – such as polarity, metabolic stability, target specificity, and hydrophobicity – and can make or break therapeutic success in drug development. In this article, Woo et al., report a novel chemical transformation, enabling direct conversion of a heteroaromatic carbon into a nitrogen atom, with the ability to quickly convert quinolines (a heterocyclic aromatic ring compound with one N) into quinazolines (an identical structure, with another C replaced by an N). This one-pot technique will expand the medicinal chemistry armamentarium and has the potential to significantly influence pharmaceutical development.Organic synthesis is challenging; although individual atom modifications would be ideal, chemists often have to manipulate a number of atoms in tandem, through a series of chemistries. The ability to convert one atom in a molecular skeleton to another has, to date, remained elusive. Woo et al., introduce a one-pot reaction to replace the carbon atom in quinolone molecules with a nitrogen atom. This necessitated the carbon atom being activated as a leaving group, with said intermediate being electrophilic to facilitate incorporation of an added nitrogen. First, the compound undergoes oxidative cleavage, and is irradiated with a 390 nm LED. Then, an ammonia nucleophile is added, before undergoing ozonolysis. The authors found that a number of quinazolines could be synthesized at high yield, and that their technique can modify carbon skeletons inside complex therapeutic molecules. This work lays the foundation for subsequent efforts for direct C → N substitutions, and this method will certainly influence medicinal chemistry for the foreseeable future.
Notable Deals
Beam Therapeutics: developing a fully integrated platform for precision genetic medicines, anchored by a key proprietary base editing approach.
Verve Therapeutics: applies gene editing to develop one-time medicines for cardiovascular diseases (CVD). Verve has an exclusive license to Beam’s base-editing technologies in 4 CVD targets.
Eli Lilly: Indianapolis pharma giant with investments in gene / base editing technologies and a pre-existing collaboration with Verve.
Deal Overview:
Eli Lilly has initiated a transaction for Beam’s stake in Verve Therapeutics. Lilly will pay Beam $250M in combined upfront payment and equity investment to acquire it’s product rights to 4 programmes in Verve's cardiovascular gene editing portfolio, plus up to $350M in development milestones. As a result, Lilly will gain a 33% cost share and 50% profit share arrangement on these programmes.
Beam Rationale:
Immediate non-dilutive cash infusion is well-timed for Beam following its recent 20% workforce reduction, extending it’s runway into H2:26. This is important for Beam to reach critical value inflection points across its portfolio.
Financial terms are attractive for Beam (and lilely a premium) given the total size of the deal ($600M) is the size of Verve’s market cap. Perhaps Lilly / Beam have had a sneak peek at Verve’s next big data readout and this data may not be as strong as hoped for.
Gate Bioscience launches with $60M and a novel small molecule play
Company: Gate Bio is building a novel class of small molecules called Molecular Gates to tackle extracellular disease causing proteins. Most small molecule therapies target intracellular proteins. The technology is based on work by Pat Sharp, a co-founder.
Technology: Molecular Gates prevent the secretion of these disease-causing proteins from the cell, preventing them from having the chance to cause disease. They do this by binding to a translocon or passage-way from inside to outside the cell.
Investors: Versant Ventures, Arch Ventures, a16z and GV
Suggested indications: inflammatory diseases, diseases caused by protein aggregation, and CNS diseases, that are not amenable to traditional biologics.
Investor sentiment:
Although the biotech public markets are bearish, biotech investors are still closing large funds to catalyse the new wave of start-ups:
Bioluminescence: $477M fund launch to invest in breakthrough biology platforms for drug discovery.
Abingworth: $356M to invest in clinical-stage biotech
OrbiMed: $4.3B fund to invest in a wide-range of innovative and growth-oriented opportunities across healthcare
Sofinnova: $200M to invest in computational biology tools, health software platforms an digital medicines.
Revelation Partners: $608M to continue offering cash in exchange for minority equity interests in private healthcare companies. This mechanism is useful for founds and VCs who need a cash injection as they await an M&A or IPO.
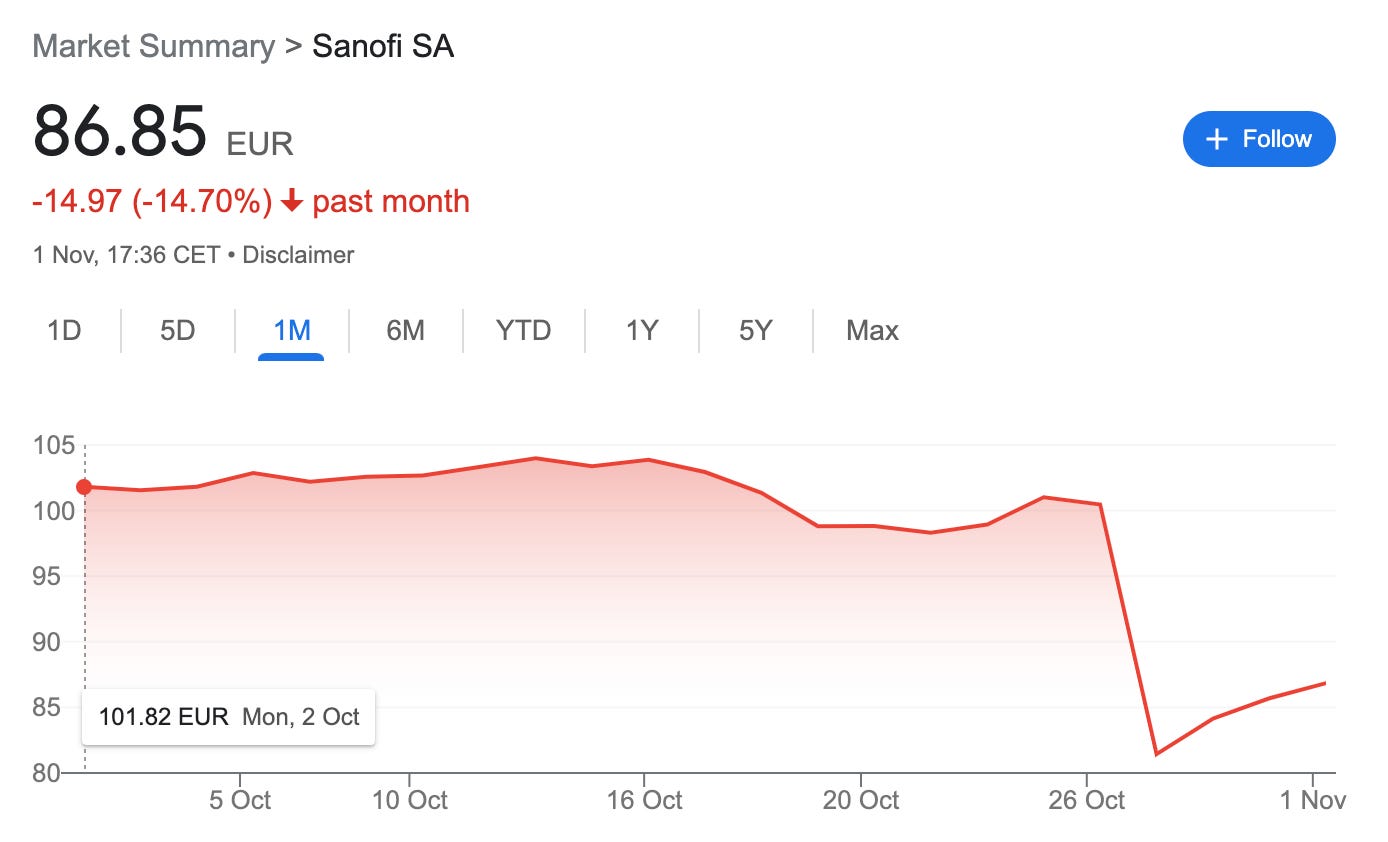
Sanofi: a market recoil
Sanofi lost approx $21B in market value post-earnings call. This may seem extreme but reflects a longstanding weariness over Dupixent-dependence and recent pipeline missteps.
What we listened to
What we liked on Twitter
Field Trip
Did we miss anything? Would you like to contribute to Decoding Bio by writing a guest post? Drop us a note here or chat with us on Twitter: @ameekapadia @ketanyerneni @morgancheatham @pablolubroth @patricksmalone