BioByte 55: a new chemistry foundation model, deep learning for affinity prediction, gliomas hijack neuronal processes, standardized single-cell data sets, Novartis calls on a Legend
Welcome to Decoding Bio, a writing collective focused on the latest scientific advancements, news, and people building at the intersection of tech x bio. If you’d like to connect or collaborate, please shoot us a note here or chat with us on Twitter: @ameekapadia @ketanyerneni @morgancheatham @pablolubroth @patricksmalone. Happy decoding!
Here’s your weekly dose of tech x bio:
Terray uses NVIDIA’s DGX Cloud to train small molecule foundation models
KDBNet: a new deep learning model for predicting binding affinity between protein kinases and drug compounds
Gliomas hijack neuronal processes, such as adaptive plasticity, which contribute towards tumor progression.
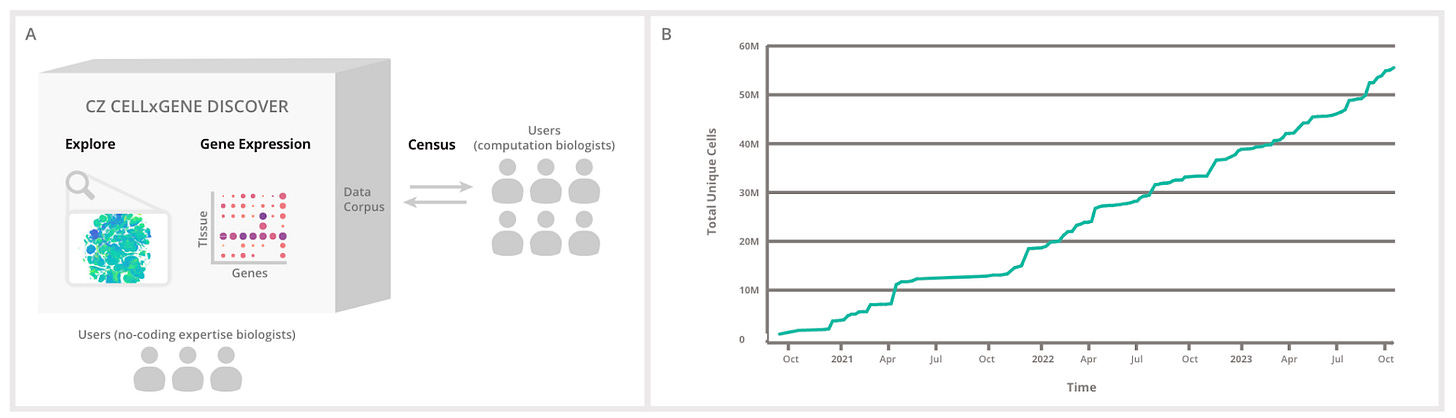
The Chan Zuckerberg Foundation released its Cell x Gene Discover platform allowing users to find and explore standardized single cell datasets.
Notable Deals: Novartis taps Legend’s CAR-Ts, Verve’s scientific breakthrough, 23&Me’s drug discovery pipeline and Turbine’s collaboration with Ono
What we read
Blogs
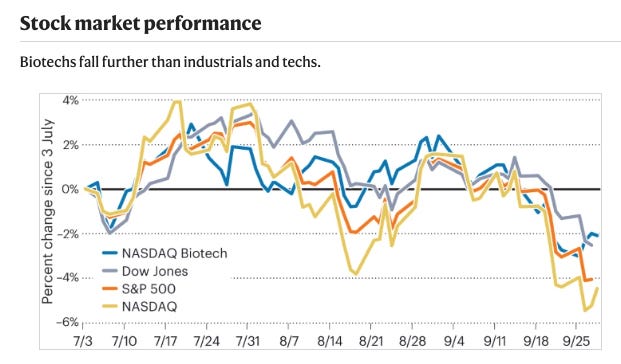
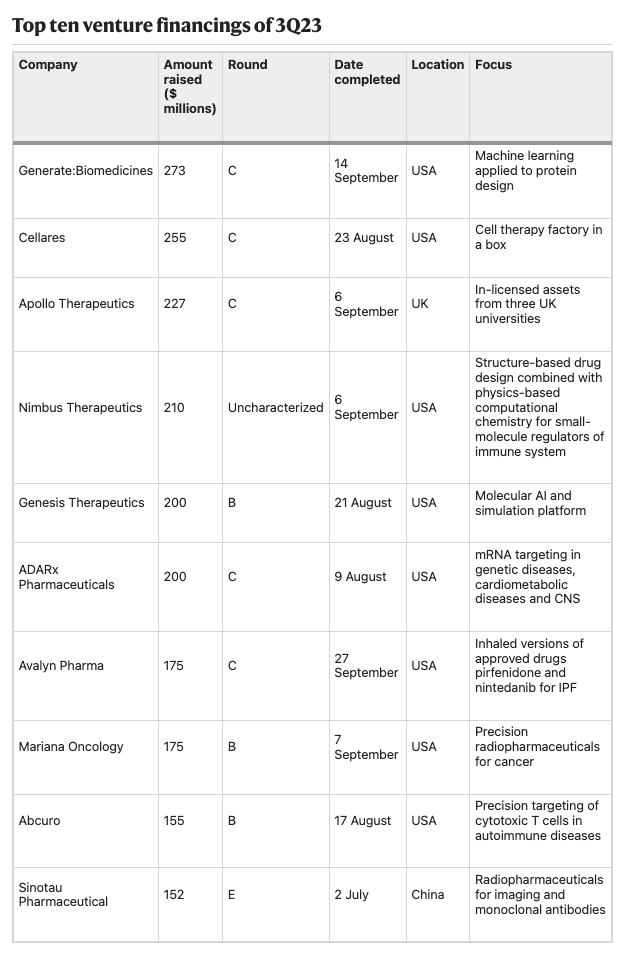
3Q23 — VCs feed ‘older children’ as IPO markets stagnate [Nature Biotechnology, Hodgson, 2023]
Insightful market summary highlighting the current state of the biotech IPO market. Overall, the biotech IPO market remains challenging, with only 7 US IPOs in Q3 2023 and just Apogee Therapeutics maintaining a steady stock price post-IPO. With few IPO options, venture financing focused on later stage companies, with 6 of the top 10 rounds being Series C. The largest was Generate Biomedicines' $273M raise. M&A provided an exit alternative, with Novo Nordisk acquiring Inversago Pharma and Embark Biotech for obesity/appetite suppression, and Danaher purchasing Abcam.
Overall, biotech stocks underperformed broader markets. Licensings and collaborations provided another funding source, with RNAi, gene therapy, and antibody technologies remaining prominent. Notable deals included Alnylam's $310M RNAi pact with Roche and Biogen's $7.3B acquisition of Reata's Friedreich's ataxia drug. While public markets lag, biotech financing continues through creative means.
A New Molecular Language for Generative AI in Small-Molecule Drug Discovery [NVIDIA, Nov 2023]
NVIDIA published a case study detailing how Terray Therapeutics (featured in the Decoding Bio 2023 Snapshot) is using NVIDIA’s DGX Cloud to train foundation models for small molecule chemistry. To efficiently explore small molecule chemical space (10^60 - or novemdecillion - possible drug-like molecules) computationally, the platform measures hundreds of millions of interactions between small molecules and biological targets daily. This exponentially growing dataset feeds into a new schema for representing small molecules called COATI, a multimodal encoder-decoder model for chemical space.
An encoder-decoder model is a type of deep learning model commonly used in NLP that involves transforming input data into output data. The encoder processes input data (in this case, small molecules represented as character strings) and converts them into a set of feature vectors called embeddings, or lower dimensional representations that capture the relevant properties of the input data. Embeddings are far more computationally efficient to optimize with AI, and therefore allow Terray to navigate chemical space to generate new small molecules with the desired drug-like properties. Once an ideal area of optimization or embedding space is identified, the decoder is used to transform an embedding back into the native space of the input data, in this case a small molecule.
Given the size of the datasets and models that Terray works with, model training became a bottleneck. Distributed training, the process of training a model across multiple computational resources to accelerate training time, can be a huge pain. NVIDIA’s DGX Cloud helped facilitate and automate distributed training. Impressively, training time for a model decreased from 1 week to 1 day.
Base editing, a new form of gene therapy, sharply lowers bad cholesterol in clinical trial [Jocelyn Kaiser, Science, November 2023]
The article discusses Verve Therapeutic’s new CRISPR strategy which, if clinical results hold, could help curb heart disease in one treatment. The new CRISPR technique involves base editing of the PCSK9 gene in the liver, which can help lower “bad” cholesterol (LDL) levels in patients with a genetic predisposition for high cholesterol. This approach is said to be safer than traditional CRISPR methods due to its targeted editing that does not involve breaking both DNA strands and lower risk of off-target effects. The CRISPR variant itself was delivered to the liver in lipid nanoparticles. The Verve trial involved 10 participants, almost all of whom had a heart attack or cardiac arrest and were born with gene mutations resulting in high cholesterol. The long-term safety effects are unstudied but in the short term, we know the CRISPR treatment is seemingly working.The next iteration of trials will involve 40 patients with familial hypercholesterolemia. Additional roadblocks in the future will inevitably involve discussions on the high costs of such a treatment.
Hey Siri: Update My Genes [Jason Steiner, Techbio<>Biotech, Nov 2023]
Jason Steiner recently republished his article on ethics and genomics from 2019, which nearly four years later is just as sharp, maybe even more realistic now. The article follows his thoughts on how environment and culture affect our genetic composition and selection, individually and from an evolutionary standpoint. Given the power of genetic engineering (see CRISPR trial covered above), it’s a timely visit to the topic of ethics and genetics. Some highlights:
This discussion and debate on ethics & genetics is not new—in many ways is a natural continuation of society’s concerns when Genentech was launched
Large-scale implications of genomics and genomic engineering is simply not well understood
Environmental and cultural influences continually shape our genetics—ex: consuming animal milk → adult lactase expression
We fear direct genetic engineering but don’t give much thought to indirect activities that end up influencing our genetics
We have a limited understanding of how organs and cellular systems interact with other another yet think we can “play god” with genomic manipulation
But what does playing god really mean? We think of it as influencing nature but you can argue the nurture part of nature vs nurture also plays a role given the strong environmental influence of genetics
Academic papers
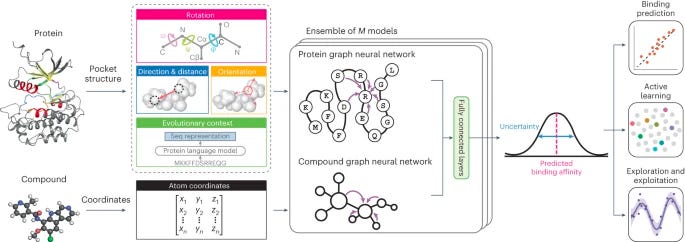
Calibrated geometric deep learning improves kinase–drug binding predictions [Luo et al., Nature Machine Intelligence, 2023]
Why it matters: This study demonstrates the value of integrating 3D structure data with deep learning for more accurate prediction of biomolecular interactions. The ability to quantify prediction uncertainty also makes the model useful for accelerating data-driven drug discovery. By better predicting binding profiles of kinase inhibitors, KDBNet can help uncover new drug candidates or repurpose existing drugs while reducing the need for exhaustive lab screening.Luo, Liu, and Peng present KDBNet, a new deep learning model for predicting binding affinity between protein kinases and drug compounds. KDBNet incorporates 3D structure information of both the kinase binding pocket and drug molecule into graph neural networks, allowing the model to learn geometry-aware representations that capture the 3D nature of binding interactions.
Experiments on public datasets showed KDBNet achieved higher accuracy in predicting kinase-drug binding affinity compared to methods using only 1D or 2D features. A key advantage of KDBNet is its ability to provide uncertainty estimates along with binding affinity predictions. The uncertainty proved useful for guiding active learning and Bayesian optimization to efficiently explore and exploit strong kinase-drug binding pairs.
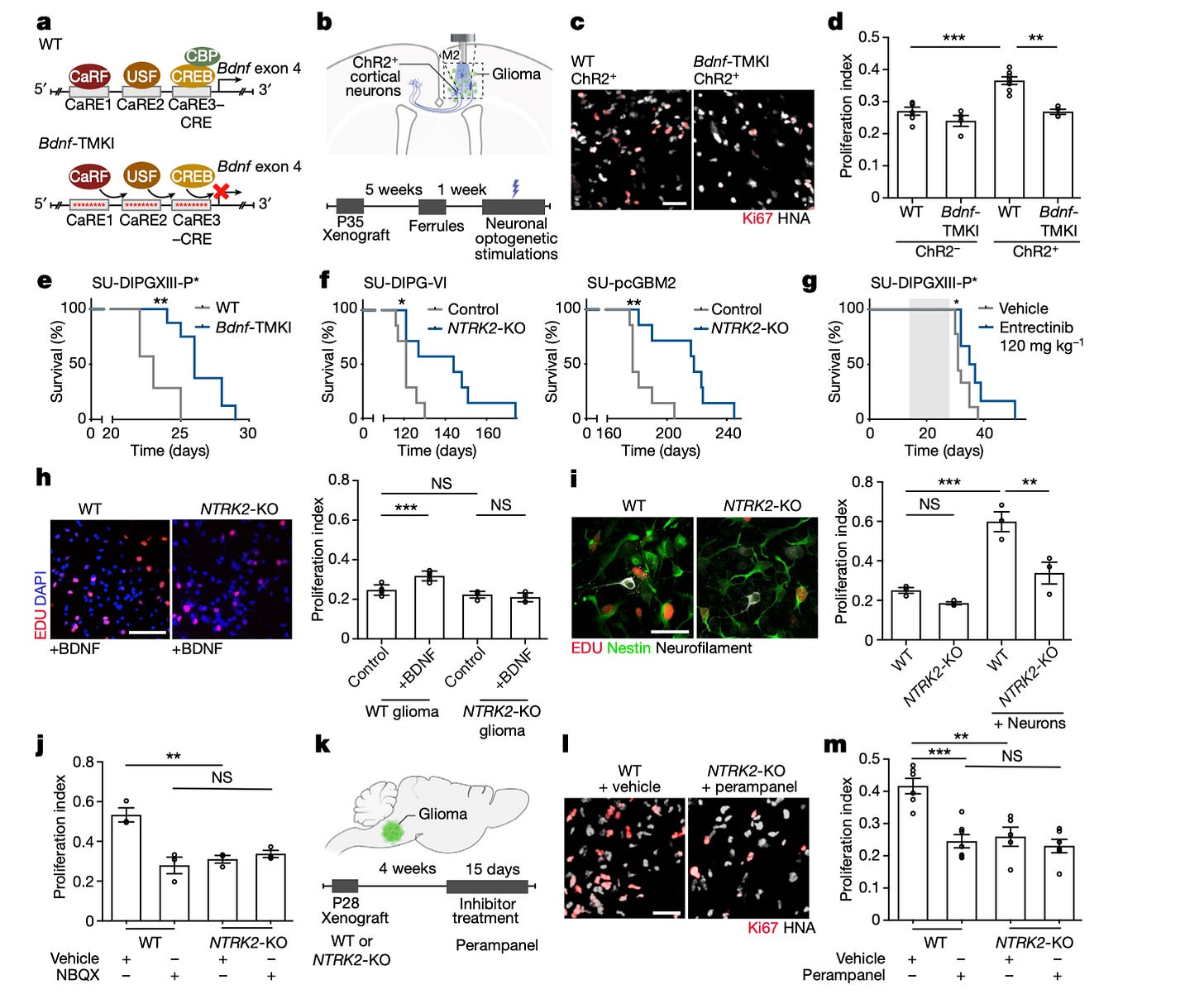
Glioma synapses recruit mechanisms of adaptive plasticity [Taylor et al., Nature, 2023]
Why it matters: Gliomas are the most common and lethal primary brain cancers, with little to no therapies available because of their inherent complexity. This paper builds on an emerging body of work probing the glioma-neuronal interaction; here, the authors demonstrate that gliomas hijack neuronal processes, such as adaptive plasticity, which contribute towards tumor progression. Gliomas share common tumor growth mechanisms with other cancers, but recent findings highlight that interactions with neurons play a crucial role in their progression and malignancy. This interaction can occur in various forms, such as paracrine signaling, leading to gliomas integrating into neural circuits. The research presented in this paper shows that gliomas can form synapses with neurons. When these neurons release glutamate, it increases the production of BDNF, a protein that binds to the TRKB receptor. This interaction boosts the expression of AMPAR, a protein on glioma cells that facilitates calcium flow and increases synaptic activity. By inhibiting TRKB or interfering with the BDNF pathway, the researchers managed to reduce cancer cell survival. This discovery lays the groundwork for potentially using TRK inhibitors, which disrupt the BDNF-TrkB pathway, in treating gliomas and opens new avenues for targeted therapies.
The APOE-R136S mutation protects against APOE4-driven Tau pathology, neurodegeneration and neuroinflammation [Nelson et al., Nature Neuro, Nov 2023]
Why it matters: A new study from Yadong Huang’s lab at the Gladstone Institute uncovers the functional role of a recently discovered APOE4 variant that protects against Alzheimer’s disease, providing a novel target for therapeutic development. Apolipoprotein E4 (APOE4) allele is the most significant genetic risk factor for Alzheimer’s disease (AD). Recently, a variant of APOE was discovered APOE3-R136S (APOE3-Christchurch) that protects against early-onset AD. The authors generated tauopathy mouse and human iPSC-derived neuron models carrying APOE4 with the homozygous or heterozygous R136S mutation, and found that homozygous R136S mutation successfully countered APOE4-driven tau pathology, neurodegeneration, and neuroinflammation. Conversely, the heterozygous R136S mutation provided partial protection against neurodegeneration and neuroinflammation caused by APOE4 but did not significantly impact tau pathology. One proposed mechanism by which the R136S mutation protects against the development of AD is reducing tau uptake in human neurons (one of the hallmark pathologies in AD leading to neurodegeneration). The mutation results in a charged variant of APOE4, which interferes with APOE4 binding of the heparin sulfate proteoglycan receptor that mediates neuronal cell tau uptake.
CZ CELL×GENE Discover: A single-cell data platform for scalable exploration, analysis and modeling of aggregated data [CZI Single-Cell Biology Program, bioRxiv, November 2023]
Why it matters: Biological data standardization and interoperability is a major issue in the life sciences, increasingly so due to the large dataset requirement to build deep learning models. In order to use cell-level data, scientists require metadata that tags a sample with important information and context about it and how the experiment was run. Only 25% of publicly available datasets provide the necessary metadata required for reuse. The CZF has released its Cell x Gene Discover platform allowing users to find and explore standardized single cell datasets.Over the past decade, research communities have aimed to clarify the molecular nature of cells, and is now more at reach than it ever has been due to the advances in single-cell technology. Various cell atlases have led to a swathe of data describing the variance across organisms, tissues, sex and ancestries, amongst others. This progress raises several issues: inconsistencies in storage formats and metadata capture.
Data standardization and interoperability is required if researchers are to use multiple data sources in a single analysis. The Chan Zuckerberg Foundation released its open source CZ Cell x Gene Discover platform, which enables scientists to “find, download, explore, analyze, and publish standardized single-cell datasets” through its no-code UX/UI and APIs.
According to the authors, the platform has already been successful:
“Researchers using the platform recently uncovered the mechanism by which aging can drive B-cell lymphoma and the identified signaling gene sets involved in small cell lung cancer. The standardized data from CellxGene has enabled the development of new computational tools including UniCell: Deconvolve Base (UCDBase), a pre-trained, interpretable, deep learning model to deconvolve cell type fractions and predict cell identity across transcriptomic datasets;67 scTab, a cell type prediction model; and CSeQTL, a tool for mapping cell type-specific gene expression quantitative trait loci”
Notable Deals
Novartis calls on a Legend for $1.1B
Legend Biotech: Commercial-stage developer of a range of CAR therapies, including the recently approved Carvykti (with J&J).
Overview: Novartis will pay $100M upfront, up to $1.01B in milestones and tiered royalties for an exclusive global license to CAR-T cell therapies targeting DLL3, including LB2102 (autologous CAR-T candidate for the treatment of SCLC and LCNEC). Novartis will fund Legend to conduct its Phase I trial and will then commence further development.
Legend rationale: Enables Legend to monetise future value streams much earlier in development to help propel company activities in a non-dilutive manner (Phase I for LB2102 will not be complete until 2028).
Investor sentiment: Provides further validation of the platform but most of Legend’s valuation still rests on Carvykti performance.
Novartis rationale: NVS is one of the CAR-T pioneers (Kymriah). It has invested in a platform to develop next generation CAR-T therapies (T-Charge) that reduces ex vivo culture and manufacturing time and is eager to cement itself in the space as per a its new ‘pure-play’ strategy.
Verve: A scientific breakthrough and a 40% stock drop
Verve presented the first results of a one-time in vivo base editing therapy in humans. Data in 10 difficult to treat patients (uncontrolled LDL familial hypercholesterolaemia) showed a marked reduction in LDL. However, a clear market and patient need is most critical to investors. Given the safety issues (one patient died of heart attack), Novartis’ twice-per-year gene silencing therapy alternative and the projected high costs clearly led investors to worry.
Recursion / Tempus: Big data gets bigger
Recursion has entered into a licensing agreement with Tempus for preferred access to its oncology-focused clinical and DNA/RNA molecular observational datasets.
Terms: Recursion will make annual payments to Tempus in cash or equity ranging between $22-42M each year, up to $160M total, over the next 5 years in exchange for continued and updated data access and use rights for therapeutic development purposes
Rationale: It is a truth generally acknowledged in ML that the bigger and more diverse a dataset, the more / interesting insights can be mined. When combined with Recursion’s proprietary dataset of over 25 petabytes of interventional biological and chemical data, Recursion will now have approximately 50 petabytes of proprietary data to facilitate the discovery of novel associations and mechanisms not otherwise identifiable in either dataset alone.
23&Me: Everyone becomes a biotech in the end
The prominent consumer genetics company has cemented itself as a serious biotech player. 23&Me sits on a treasure trove of patient data (14M patients compared to 500K of the UK biobank) of which it can mine novel insights to inform drug discovery. Purely selling insights to pharma to develop drugs leaves a lot of value on the table and so 23&me (as with other ‘platform’ biotechs) have sought to create their own therapeutics. 23&me has 100 drug hunters, an eminent CSO, a large collaboration with GSK and two IO candidates in the clinic.
Turbine.ai has signed a collaboration with Japanese pharma Ono to use its computational cell platform to find novel first-in-class targets. Turbine will conduct in vitro and in vivo validation of identified targets, which Ono will then have the right to license for a fee and future milestones. Ono is taking bet on an interesting new technology and pays for success. Ono already has partnerships with Bayer and Cancer Research UK.
In case you missed it
Hallucinating hallucinogens [Skinnider, Science, November 2023]
From the Decoding Bio Team:
The Faster Horse Problem in AI [Morgan Cheatham, Decoding Bio Contributor]
On biotech platform strategy [Patrick Malone, Decoding Bio Contributor]
What we liked on social channels
Field Trip
Did we miss anything? Would you like to contribute to Decoding Bio by writing a guest post? Drop us a note here or chat with us on Twitter: @ameekapadia @ketanyerneni @morgancheatham @pablolubroth @patricksmalone