BioByte 003
Mind reading using ML, The Era of Cheap Genome Sequencing, A Positive Amyloid Trial, Digital Scent, FDA Modernization Act, and Space Skeletons?
Welcome to Decoding TechBio, a writing collective focused on the latest scientific advancements, news, and people building at the intersection of tech x bio. Subscribe for free to read our weekly review and deep dives on areas we’re excited about. For more on us, check out our mission statement here. If you’d like to connect or collaborate, please shoot us a note at decodingtechbio@gmail.com. Happy decoding!
“CRISPR in the style of Kandinsky” in homage to OpenAI releasing its beta to the public this week
What a week! The last seven days were among some of the most transformative for the field of biology (we feel this way many weeks, tbh): starting with a positive amyloid trial for the treatment of Alzheimer’s, Illumina announcing its $200 whole genome sequencer, and the FDA passing a bill that removes the mandate for animal data in drug development. This is a longer post for us, but we break it all down below.
What we read
Academic Papers
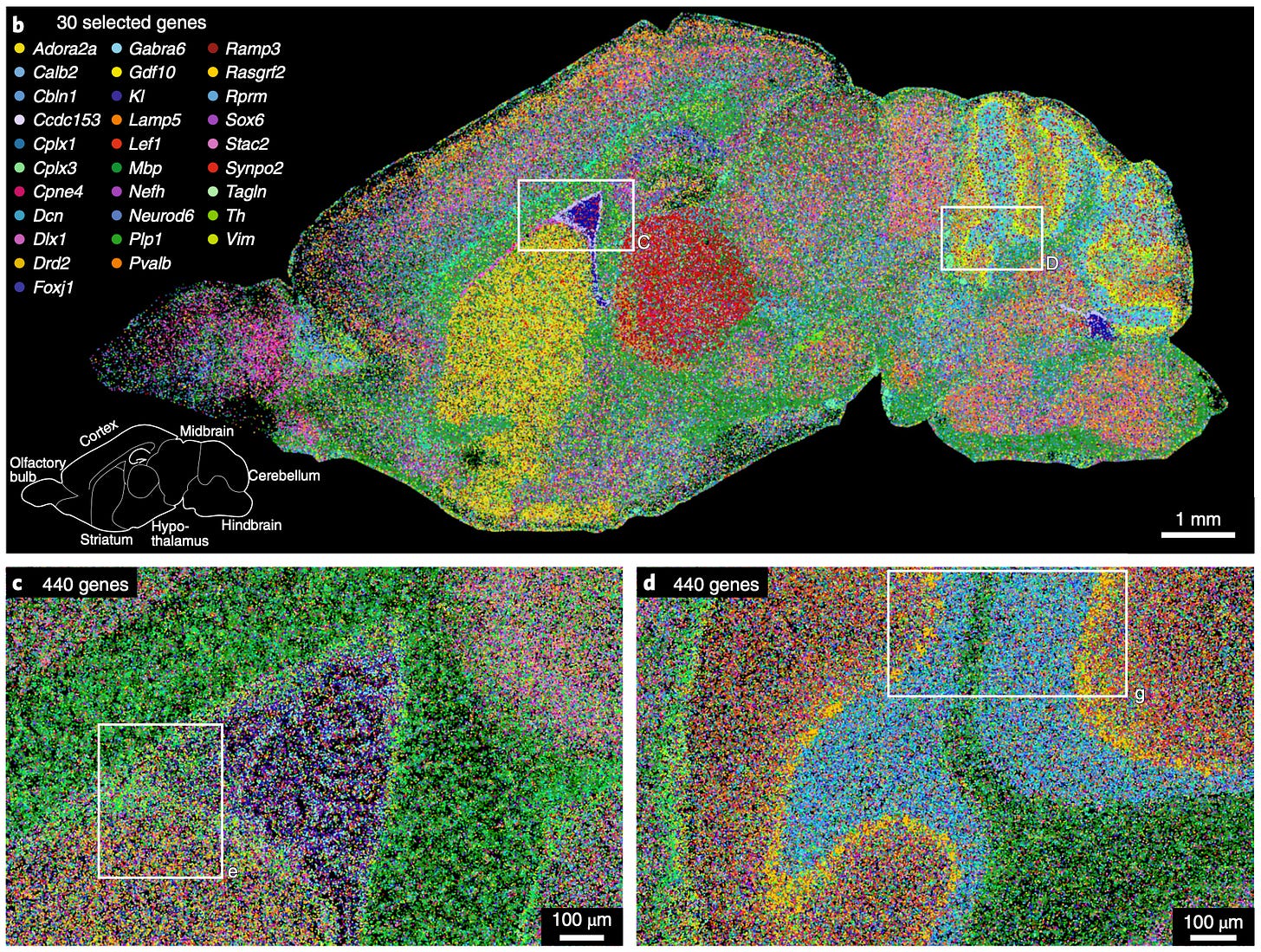
Scalable in situ single-cell profiling by electrophoretic capture of mRNA using EEL FISH [Born et al., Nature Biotech, 2022]
Single-cell spatial transcriptomic profiling has enabled the classification of cellular populations with remarkable resolution. This has been especially important in organs that have vast cellular diversity like the brain. However, current methods, via sequencing or microscopy, are dominated by a trade-off between resolution and throughput. The authors present a method named Enhanced ELectric Fluorescence in situ Hybridization (EEL FISH), that can rapidly process large tissue samples without compromising spatial resolution. They build from both microscopy and single-molecule fluorescence in situ hybridization (smFISH) techniques to solve for the drawbacks of each method when used individually.
Pan-cancer analyses reveal cancer-type specific fungal ecologies and bacterium interactions. [Narunsky-Haziza et al., Cell 2022]
The tumor microenvironment is its own unique ecosystem. Although the tumor-bacteria relationship has been well explored, the role of fungi and its interplay with cancer has been poorly understood. Here, the authors develop the "Pan-Cancer Mycobiome Atlas” and found that different cancer types exhibit cancer-type-specific mycobiome profiles, and that the presence of intratumoral fungi could be used to stratify clinical outcomes. Additionally, ML models assessing cell-free fungal DNA could classify healthy vs cancer patients.
It’s a microbial world, and we’re all just living in it…
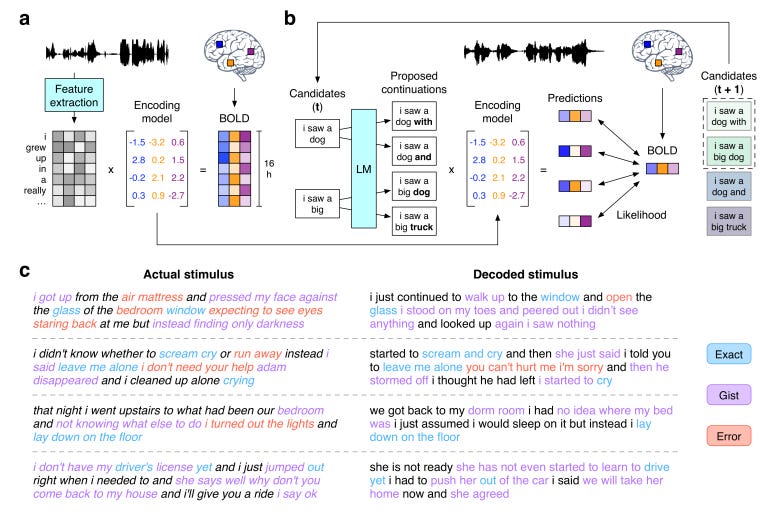
Semantic reconstruction of continuous language from non-invasive brain recordings [Tang et al., BioRxiv 2022]
The field of neural decoding, or reading out mental activity by applying machine learning to neuroimaging data like functional MRI or EEG, has exploded in recent years. In this paper, Alex Huth’s lab analyzed fMRI data acquired while participants listened to spoken stories. Using a bayesian decoding algorithm that combines language models and encoding models (commonly used in neural decoding of fMRI data), the authors could reconstruct the meaning of perceived speech on the basis of the fMRI data alone. The field of neural decoding has grappled with questions of privacy; does this technology compromise mental privacy, exposing our inner thoughts to intruders without our permission? Tang et al., tested a few mitigation strategies, such as performing a mental calculation, and showed that neural decoding performance was decreased.
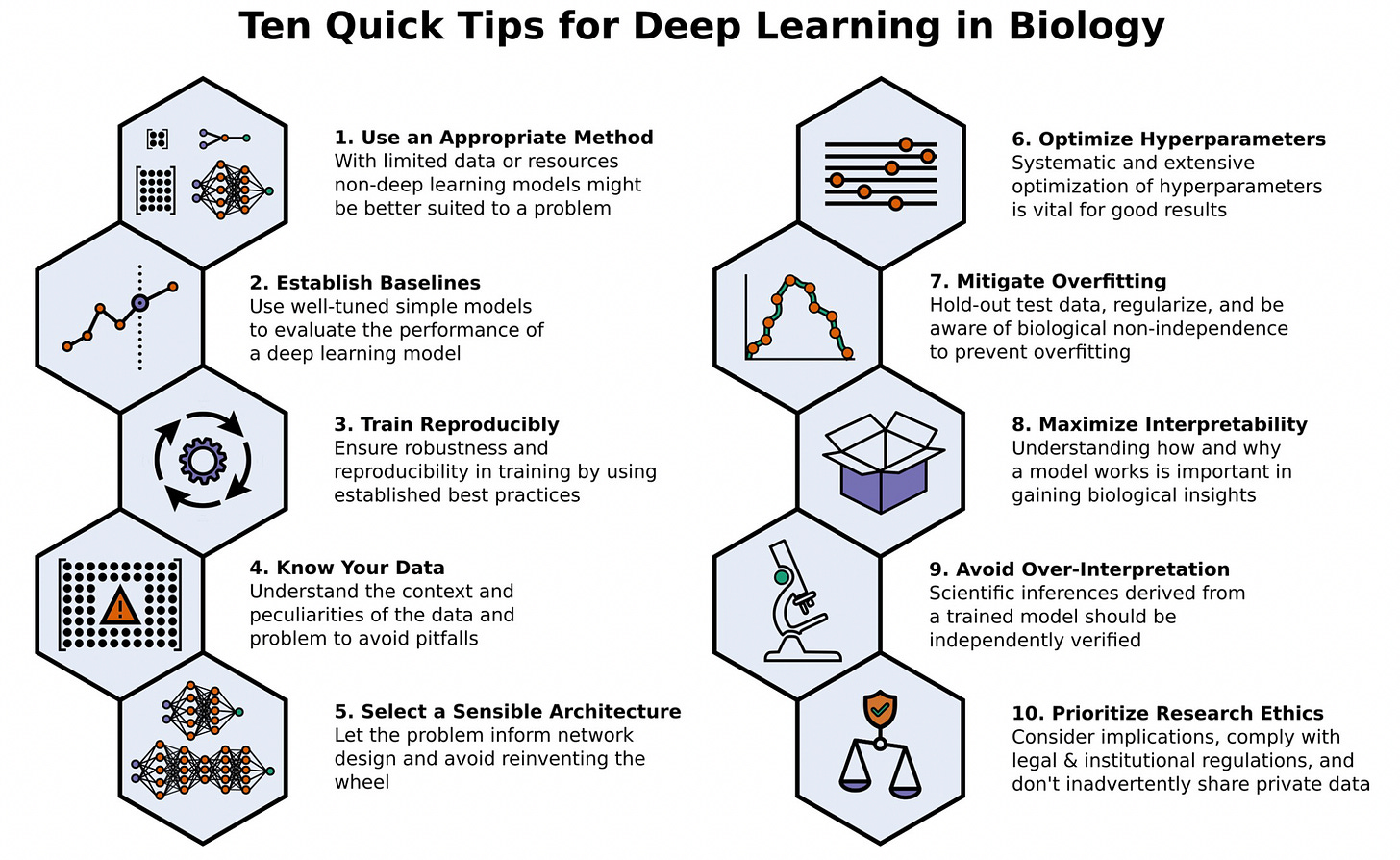
Ten quick tips for deep learning in biology [Lee et al., PLOS Comp Bio 2021]
Lucid overview of practical tips for applying deep learning in biology. The paper is wroth a read, but one of our favorite tips:
Establish a baseline performance with a simpler, linear model. Only use more complex, non-linear models when necessary. For startup founders, avoid using deep learning simply because it might generate more VC interest. If you can derive similar performance from simpler models, brag about it! Simpler models are faster and cheaper to train, are more interpretable, and you won’t have to pay a 23-year old ML engineer 300k/year.
Controlling gene expression with deep generative design of regulatory DNA [Zrimec et al., Nature 2022]
Advancements in generative artificial intelligence are taking the world by storm from GPT-3 to DALLE, and biology is not immune. This recent paper prototyped a deep learning model using generative adversarial networks (GANs) to design de novo synthetic regulatory DNA, which consists of promoters, enhancers, silencers, and insulators. This work could represent a material improvement on existing methods, such as mutagenesis, that require extremely large random DNA libraries for screening. As the article highlights, shortcomings of mutagenesis are multi-fold, and include the fact that many mutagenesis-producted selection candidates are not feasible regulatory DNA, which drives inefficiencies in our search for functional sequence variants and places undue constraints on the search space. The models developed by Zrimec et al. are instead learned directly from genomic and transcriptomic data of Saccharomyces cerevisiae to generate de novo functional regulatory DNA. With >10^60 ways to construct a mere 100 bp promoter sequence, new methods for exploring this search space remain an exciting area within methods development. You can find the code for the GANs here.
Blogs
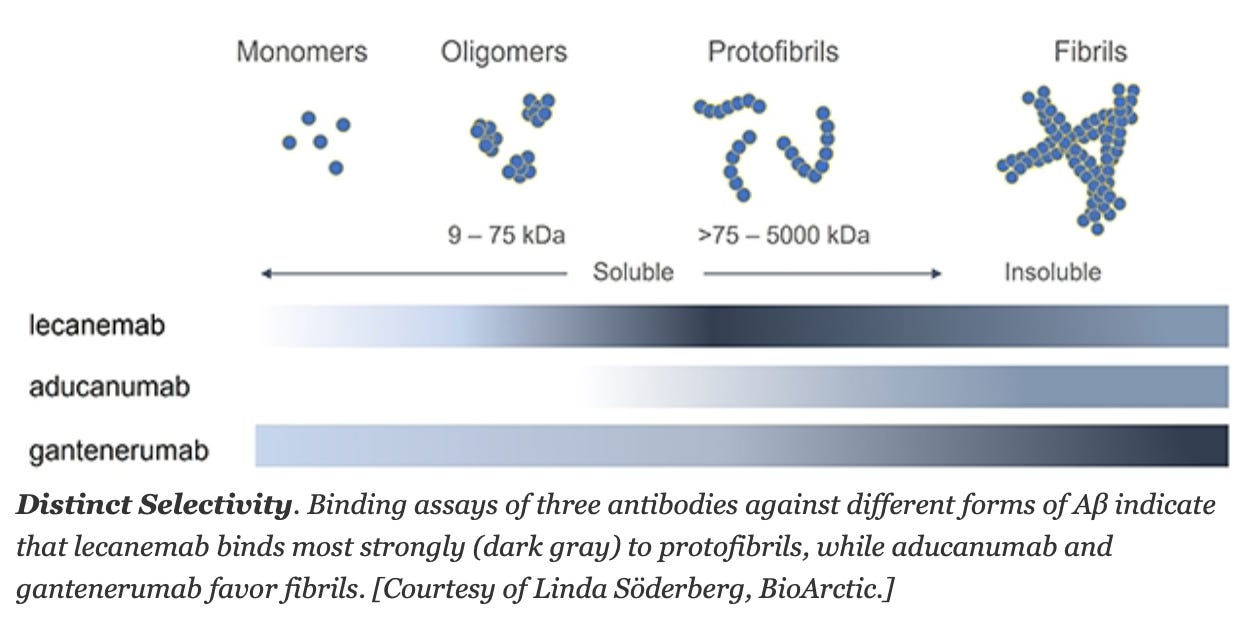
A Positive Amyloid Trial, Finally? [Derek Lowe, Science]
Big news of the week is Eisai and Biogen’s positive read out of lecanemab, which met all clinical endpoints for the treatment of Alzheimer’s. Whilst news outlets rushed with positive headlines and Biogen’s stock soared, Derek Lowe brings a different perspective to balance the argument.
There are three main concerns that he poses:
A statistically significant decrease in the primary endpoint (CDR-SB), does not equal a significant difference to the patient
The incidence of cerebral edema caused by lecanemab being too high
Alduhelm, the FDA-approved controversial anti-Abeta mAb, which ended up being a clinical and commercial flop, had two readouts one positive (a CDR-SB score of 0.39) and one that showed no such effect, where lecanemab had one read out of 0.45.
The question is, what would have happened if the companies had run a second trial? Some important questions raised that will only be answered with additional data in November.
GV aims to build a company to sniff out disease [Connie Loizos, TechCrunch]
Scent represents another layer of molecular information about the world around us, and remains largely unexplored. GV has tapped Alex Wiltschko as its newest entrepreneur-in-residence, with the goal of digitizing scent. For years, we’ve known that disease states can alter the release of volatile organic compounds from the human body – but we’ve been unable to decode and understand the information stored within. What if we could digitize scent and create the next generation of scent-based diagnostics?
"...computers can’t smell. They have no ability to detect the chemical world [so] we can’t store the really powerful memories that we associate with smell, like the smell of my grandmother’s home. It’s just gone. It only lives in my mind. The smells of people who I love, and of places I’ve been, are completely ephemeral today.
We [also] know that diseases have a smell. We know that different wellness and health states have a smell. Plants, when they’re sick or when they’re healthy, smell different. The amount of information that’s out there in the world that we could potentially act on to make our lives longer, make our lives more joyous, grow more food — that’s really only able to happen inside of living things inside of living noses — if we could automate that, we could have a massive and positive impact on society." -Alex Wiltschko, PhDThe Era of Fast, Cheap Genome Sequencing Is Here [Emily Mullin, Wired]
Ultima Genomics promised the $100 genome by 2023, Singular Genomics and Element Biosciences have developed smaller, bench top sequencing machines and MGI has started selling its sequencers; all of which will eat into the major player’s 80% share in the DNA sequencing market. How has Illumina responded? This week it announced its fastest, most cost-efficient sequencing machines yet, the NovaSeq X series. The company believes it will decrease the cost of Whole Genome Sequencing to $200 while providing a readout at twice the speed.
For its latest machines, Illumina invented denser flow cells to increase data yield and new chemical reagents, which enable faster reads of bases. “The molecules in that sequencing chemistry are much stronger. They can resist heat, they can resist water, and because they’re so much tougher, we can subject them to more laser power and can scan them faster. That’s the heart of the engine that allows us to get so much more data faster and at lower costs,” says Alex Aravanis, Illumina’s CTO
Senate passes bill to rework animal testing requirements for drug developers [Zachary Brennan, Endpts]
This week, the US Senate passed the FDA Modernization Act 2.0 via unanimous consent. The bipartisan bill alters a federal mandate for animal testing on new drugs. Within the bill, it appears the term “animal” will be replaced in several sections of the FDCA, while the term “nonclinical test” will be included. Nonclinical tests are defined as in vitro, in silico, in chemico, and other nonhuman in vivo testing that may include animal tests – a boon for TechBio-based approaches. Jeffrey Singer of the Cato Institute commented on the news expressing that though a great start, more comprehensive reform is needed.
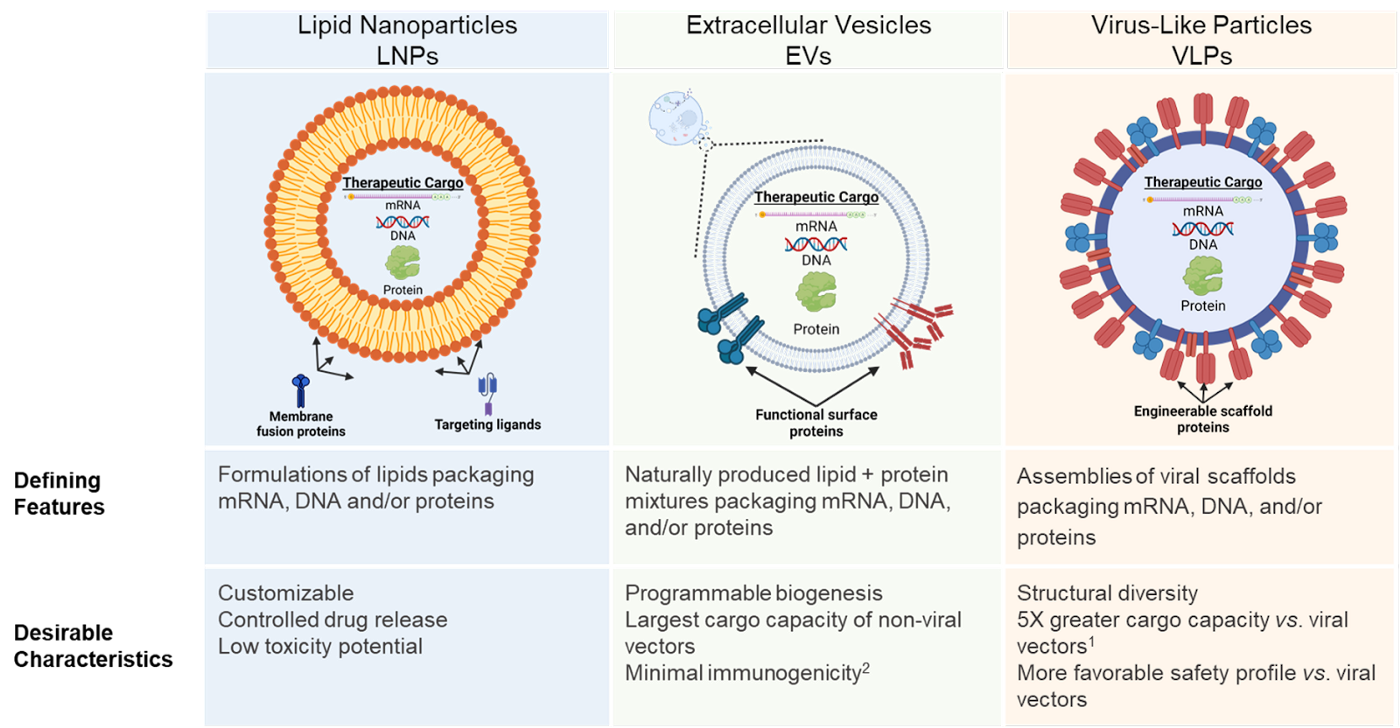
Expanding the Drug Delivery Toolbox [Emilio Ferrara and Tamara Vital, KdT Ventures]
Drug delivery is one of the most difficult challenges facing biotechnology. From Emilio Ferrara and Tamara Vital: “While today’s viral delivery methods are equipped to treat a subset of disease types, the future biotech delivery toolbox needs a more robust toolset. We believe that non-viral vectors, especially EVs, are primed to be the swiss army knife of delivery and usher in this next generation of therapies.”
The Changing World of Life Sciences R&D [Jacob Effron, Principal at Redpoint Ventures and Author of Vital Signs]
Jacob from Redpoint Ventures breaks down the current Research and Development software landscape. He explains what is happening in the field:
The next generation of vertical software tools built for life sciences companies is saving researchers time in their processes.
New infrastructure is enabling much more sophisticated and repeatable analyses of data generated in labs.
Life sciences companies are increasingly looking to AI startups to help with R&D processes.
Learning the Language of Variants [Elliot Hershberg, Century of Bio and Not Boring]
We’re big Elliot stans here at Decoding TechBio, and his latest piece on protein language models for variant prediction, and more specifically, leveraging transformer-based models to predict disease variants in silico. The piece provides an excellent distinction between the utility of AlphaFold, which helps researchers leap from sequence to structure, and recent work to go from structure to function, which will ultimately help us better understand how variants relate to pathology. This work is highly aligned with Eric Topol’s piece that we highlighted last week on Human Genomics vs. Clinical Genomics.
What we listened to
Dina Radenkovic (CEO of Gameto) interviews Josh Wolfe (Lux) and Robert Nelson (ARCH) on the future of tech x bio.
This podcast episode in Big Biology talks about model organisms – from mice, to rats, to fruit flies, and others - and both the insights gleaned and limitations of working with them. We like this podcast in the context of the passing of the FDA Modernization Act 2.0.
This Bioinformatics and Beyond Podcast seems to have paused new episodes, but this one is fantastic. Dr. Justin Siegal speaks to the past, present, and future of wet lab work and wet lab automation, and paints a picture of burgeoning “cloud lab” models. He shares his personal experience using cloud labs and how things like the accuracy and reliability of cloud labs can already make it a viable option for automating academic lab tasks.
Two heavy hitters in TechBio discuss how their companies are advancing the future of AI enabled drug discovery.
What we liked on Twitter






Where we went (and are going)
We’re excited about this upcoming session on October 19th with Nish Bhat, Co-Founder of Carbon Silicon Ventures and Color Genomics hosted by Cromatic. It looks sold out here, but maybe you can DM your way to a spot… You might even find a familiar face speaking
Private financings this week
Kaleidoscope Bio raised a $6M seed round from Hummingbird Ventures and Dimension Capital to build the connective tissue across wet lab and dry lab, making science more collaborative, reproducible, and scalable.
Form Bio raised a $30M Series A led by JAZZ Venture Partners to spin computational platform out of Colossal Biosciences. Form is building software tools to enhance bioinformatics workflows and enhance communication within the life sciences.
Cathie Wood’s ARK launches venture fund open to individual investors, for a minimum initial investment of $500. The venture fund intends to invest in early- to late-stage private companies and publicly traded businesses similar to those held by ARK’s ETF.
EnPlusOne launched from the Wyss Institute with $10M from Northpond Ventures, Breakout Ventures, and Coatue. The team is building an enzymatic synthesis platform to manufacture RNA therapeutics.
Meliora Therapeutics raised an $11M seed round to understand and catalog the mechanism of action of cancer drugs. HOF Capital and ZhenFund led the round with participation from Obvious, VillageGlobal, Pebblebed, Axial, and others.
From the community
We’re always looking for great content and feedback. If we missed anything, you’d like to collaborate, write a guest post, or just say hello, email us at decodingtechbio@gmail.com.
Field trip
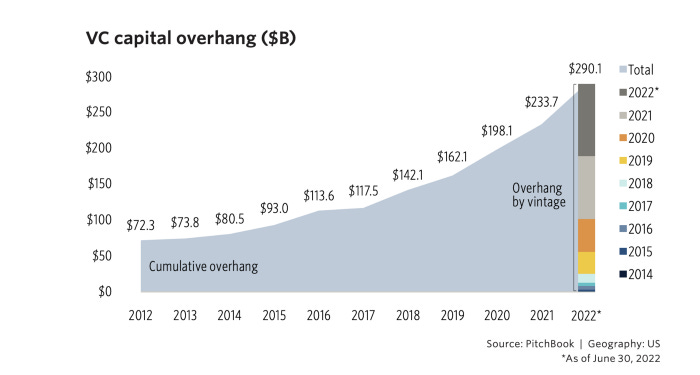
What Does the Post Crash VC Market Look Like? [Mark Suster, Both Sides of the Table]
Mark from Upront Ventures describes the new normal in valuations and expectations. Despite the $290B VC “overhang” (money waiting to be deployed into tech startups), VCs will have be patient and wait for the companies that have ‘reset’ their valuation expectations to the current market. Mark argues VCs have two options: 1) to super size or 2) to super focus.
Dall-E is now available without a waitlist
Unleash your inner artist by telling a computer what to create. Check out some renditions of “A skeleton with an electric guitar in space, playing heavy metal with sound radiating outwards, in a photorealistic style”. Not bad, not bad….
Did we miss anything? Would you like to contribute to Decoding TechBio by writing a guest post? Drop us a note at decodingtechbio@gmail.com or chat with us on Twitter: @ameekapadia @pablolubroth @patricksmalone @morgancheatham @ketanyerneni











Loved this! So glad this weekly review now exists!