BioByte 005
predictive physicochemistry, the future of synthetic biology, cultured neurons that can play pong, RNA-targeting small molecules and more
Welcome to Decoding TechBio, a writing collective focused on the latest scientific advancements, news, and people building at the intersection of tech x bio. Subscribe for free to read our weekly review and deep dives on areas we’re excited about. For more on us, check out our mission statement here. If you’d like to connect or collaborate, please shoot us a note at decodingtechbio@gmail.com. Happy decoding!
Happy Monday! We wanted to start with a quick thank you to everyone who has been sending us feedback — we’ve now split up this week’s newsletter into Content and Community and added a ‘why it’s relevant’ block from our perspective to each academic paper. We want to keep improving. If you have any feedback, shoot us an email.
What we read
Blogs
TRANSFORMing natural product drug discovery: machine learning for high-fidelity chemical property prediction from metabolomics data [August Allen, Enveda Bio, 2022]
This is the first platform of its kind to accurately predict physicochemical properties of natural compounds without using a manual reference library. This means that without testing the small molecule physically, one can determine if it is 'drug-like'. Around 60% of all FDA-approved small molecule drugs through 2020 were derived from nature. Why then don't all pharma companies focus on natural products as their starting points for drug discovery?
The answer is that analytical workflows in modern drug discovery are incompatible with natural product libraries due to their inability to:
quickly prioritize lead-like structures [“Chemical Annotation”]
confidently identify the bioactive molecule in a mixture [“Biological Annotation”]
access enough material to enable preclinical, clinical, and commercial development [“Material Access”]
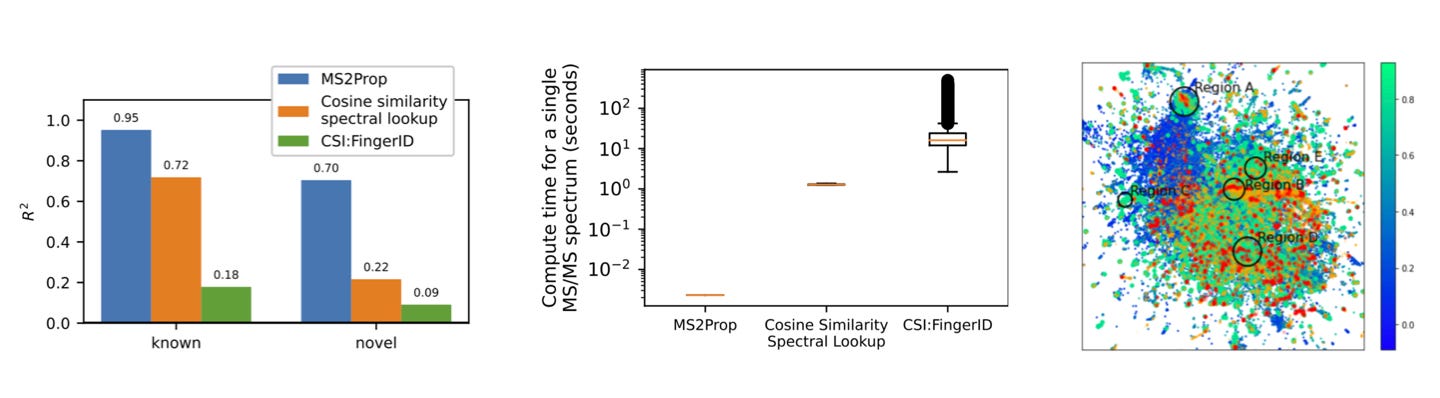
In its new publication, the Enveda Bio team discuss how they solve these issues to bring natural product drug discovery to modernity. By 'cracking' MS spectra analysis through its MS2Prop platform, the company can predict chemically relevant properties of compounds for drug discovery with an R2 of 70% (leftmost fig below). MS2Prop also enables orders of magnitude faster analysis (12,000x on average) than the state of the art (central fig), and suggests where to look for high-potential chemical space: i.e. drug-likeness regions that don't have a high concentration of FDA-approved drugs (rightmost fig).
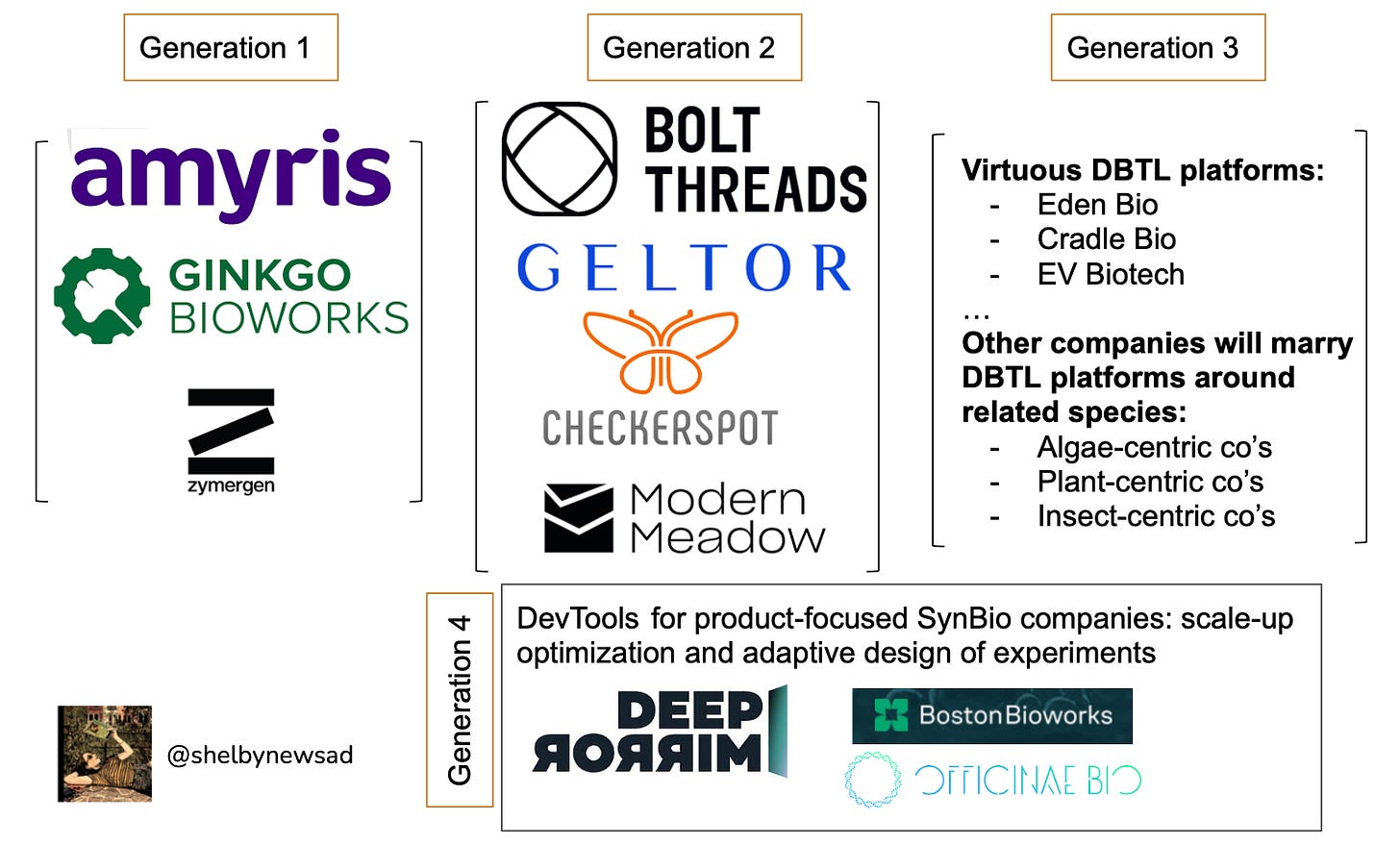
Product vs platform in synthetic biology [Shelby Newsad, 2022]
If techbio is the evolution of biotech, then synbio is techbio’s sibling. Our friend Shelby does a great job tracking the evolution of synthetic biology from synbio 1.0 of platforms to synbio 2.0 of products to synbio 3.0 of DTBL (chassis engineering, design) to synbio 4.0 of enabling scale-up infrastructure.
She argues that we have yet to see what outcomes the product-oriented companies of the last ten years will have. While many of these ingredients-as-a-service or clean chemicals companies make sense and were more de-risked from a funding perspective, the future of synbio lies in companies that can go from service —> platform with speed and proactive learning based on existing data architecture, chassis engineering, automation, etc.
Because of this, I envisage the greatest successes coming from companies and platforms built around related chassis (fungi or algae or bacteria) so they are able to capitalize on learnings and share genetic parts. With this lens, I believe a company like Phycoworks, which is using ML models to optimize product production in algae is ahead of its time. Unlike the tech industry, and as highlighted in Dennis Gong’s Anti-TechBio, biology is context dependent. Companies that work with the context dependence of biological systems will be able to build interoperable learnings and parts creating tremendous moats around their products. The next piece of the puzzle is elucidating the most versatile, fast-growing, and nimble suite of chassis on which to build.
As it is for its rote biology counterpart, in synbio, context is key.
DeSci 101 [Morgan Cheatham and Lindsey Li, Bessemer Venture Partners, 2022]
This past week, Bessemer Venture Partners published a primer on the field of decentralized science through the lens of three founders in the space: Dr. Tyler Golato of MoleculeDAO, Dr. Nikolas Rindtoff of LabDAO and Dr. Alok Tayi of Vibe Bio
We love Tyler’s articulation of the “five fundamental promises of DeSci”:
Clever people can work on anything that excites them. Anyone can fund and participate in science, independent of institution, geography, and educational background.
Scientists can spend 80% of time doing science and 20% or less applying for funding, not the other way around.
Researchers and patient communities govern cures and access to medications
We share our knowledge, data, and resources openly. Negative data is openly published and communicated.
Science decouples from institutions, and scientists are fairly paid and incentivized for their work.
Academic papers
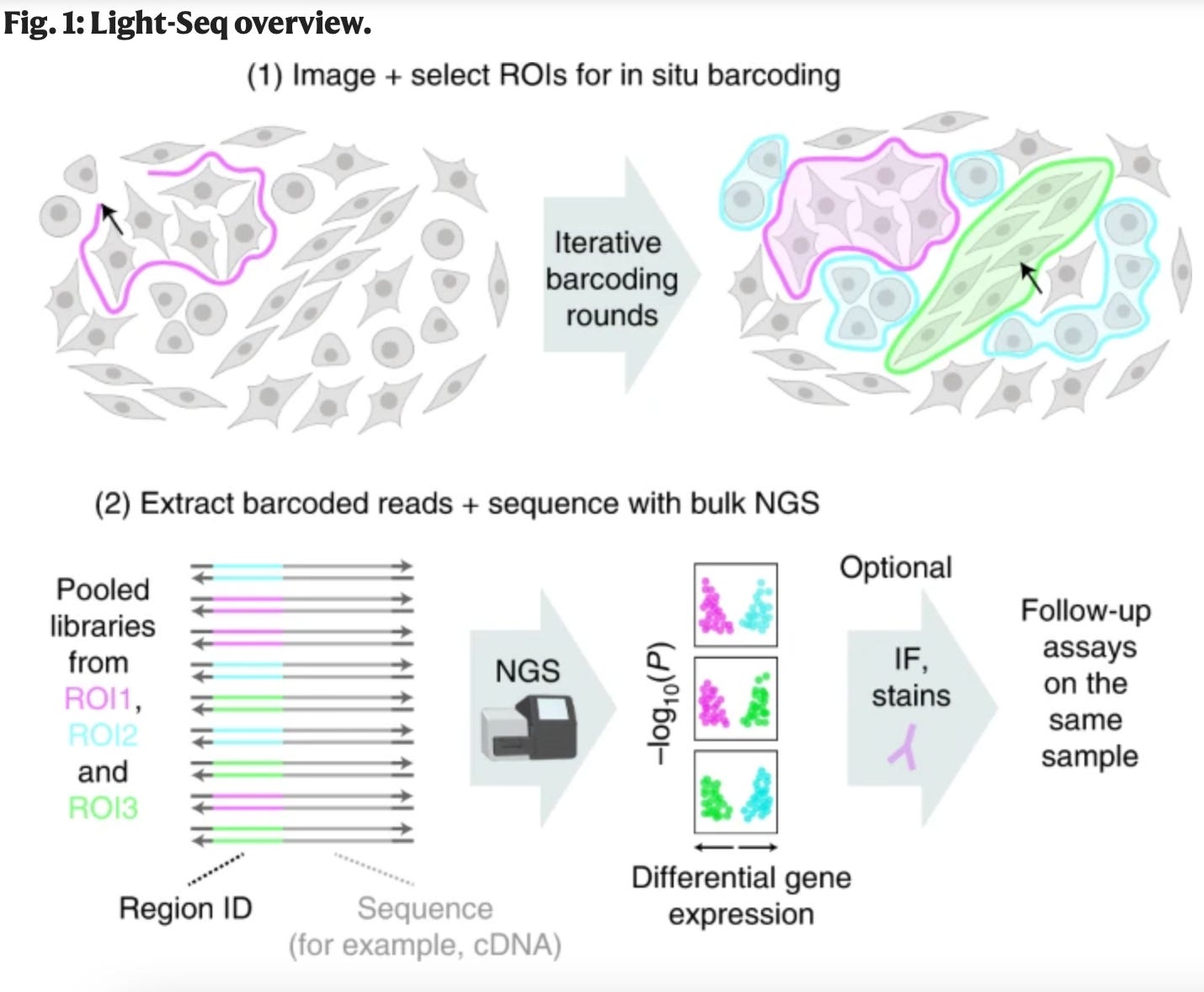
Light-Seq: light directed in situ barcoding of biomolecules in fixed cells and tissue for spatially indexed sequencing [Yin et al., Nature 2022]
What if you could image a sample, sequence specific cells from the image, and then go back to the same cells you sequenced to image again? In this Nature paper, Jocelyn Kishi, Emma West, and their colleagues combine photo-crosslinking with DNA barcoding to do just that. The process goes something like this: 1) use in situ reverse transcription to make RNA copies (cDNA), 2) focus UV light to barcode specific cells, 3) degrade the RNA to release the cDNA that is now barcoded. (interestingly, this step leaves any attached proteins and biomolecules in place for future assaying), 4) process sample with novel chemistry and sequence sample.
A more complete overview of the technology, including an animation of the process can be found here.
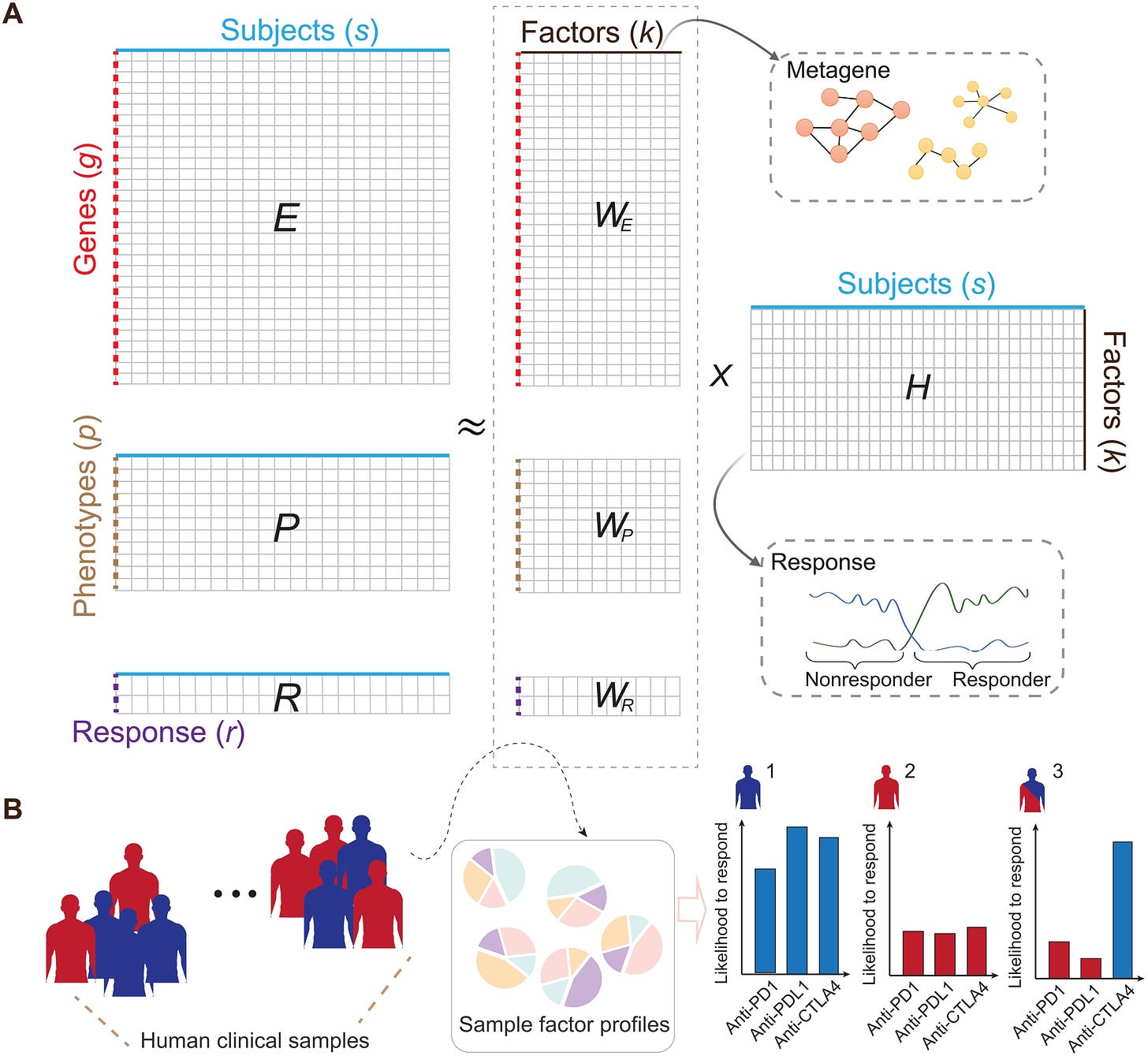
This technology is particularly exciting because it enables full transcriptome sequencing based on location and morphology without requiring cellular disruption. Researchers using Light-Seq would be able to go back to specific cell samples and run additional assays after sequencing for more comprehensive analysis. Machine learning on syngeneic mouse tumor profiles to model clinical immunotherapy response [Zeng et al., Science Advances 2022]
Immunotherapy is not the magic bullet many think it is – it can be highly toxic, and response rates are low. An open question in the field is: how can we better predict clinical immunotherapy responses? Here, Zeng et al.. develop a machine learning framework integrating transcriptomic, phenotypic, and immune checkpoint blockade (ICB) response data of 761 ICB-treated syngeneic mouse tumors from 26 published papers. They found that their model could predict clinical ICB responses and identified putative resistance mechanisms.
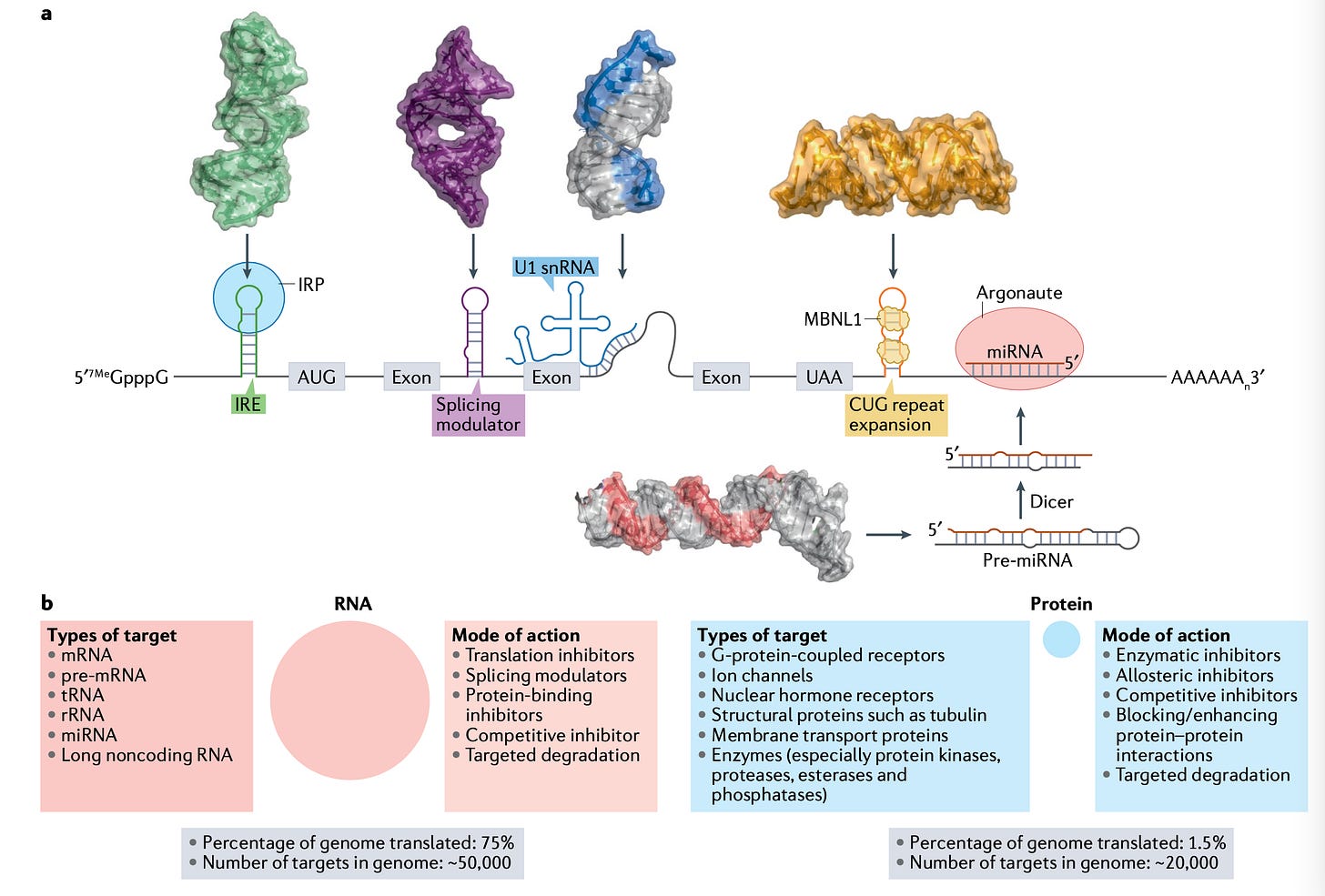
We are increasingly convinced that software-driven patient stratification will be the most efficacious and cost-effective approach to identifying the ideal patient subpopulations across all trials in medicine.Targeting RNA structures with small molecules [Childs-Disney et al., Nature Reviews Drug Discovery, 2022]
Despite the importance of RNA in disease, the advantages of small molecules and the fact that 75% of the genome is transcribed, but only 1.5% is translated, there are still tremendous challenges in drug discovery for RNA targeting small molecules. This in-depth review describes the challenges in RNA-targeting small molecule drug discovery, and the common strategies used to identify targets and lead optimise compounds to produce disease-modifying medicines.Once thought as a simple intermediate between DNA and protein, RNA is now recognised for its complex role in homeostasis and disease. Advanced therapeutics that target RNA instead of protein, such as antisense oligonucleotides (ASOs) and CRISPR gene editing, have shown great value to patients, however, translating these technologies to the clinic has been challenging owing to difficulties with delivery and significant adverse reactions. In order to overcome these challenges, researchers have developed small molecules that recognize RNA structures; which have clear advantages:
Small molecules offer an important alternative with potential for oral bioavailability and blood– brain barrier penetrance, particularly with the wealth of knowledge from medicinal chemistry whereby physico- chemical properties can be systematically optimized to improve pharmacokinetics and potency
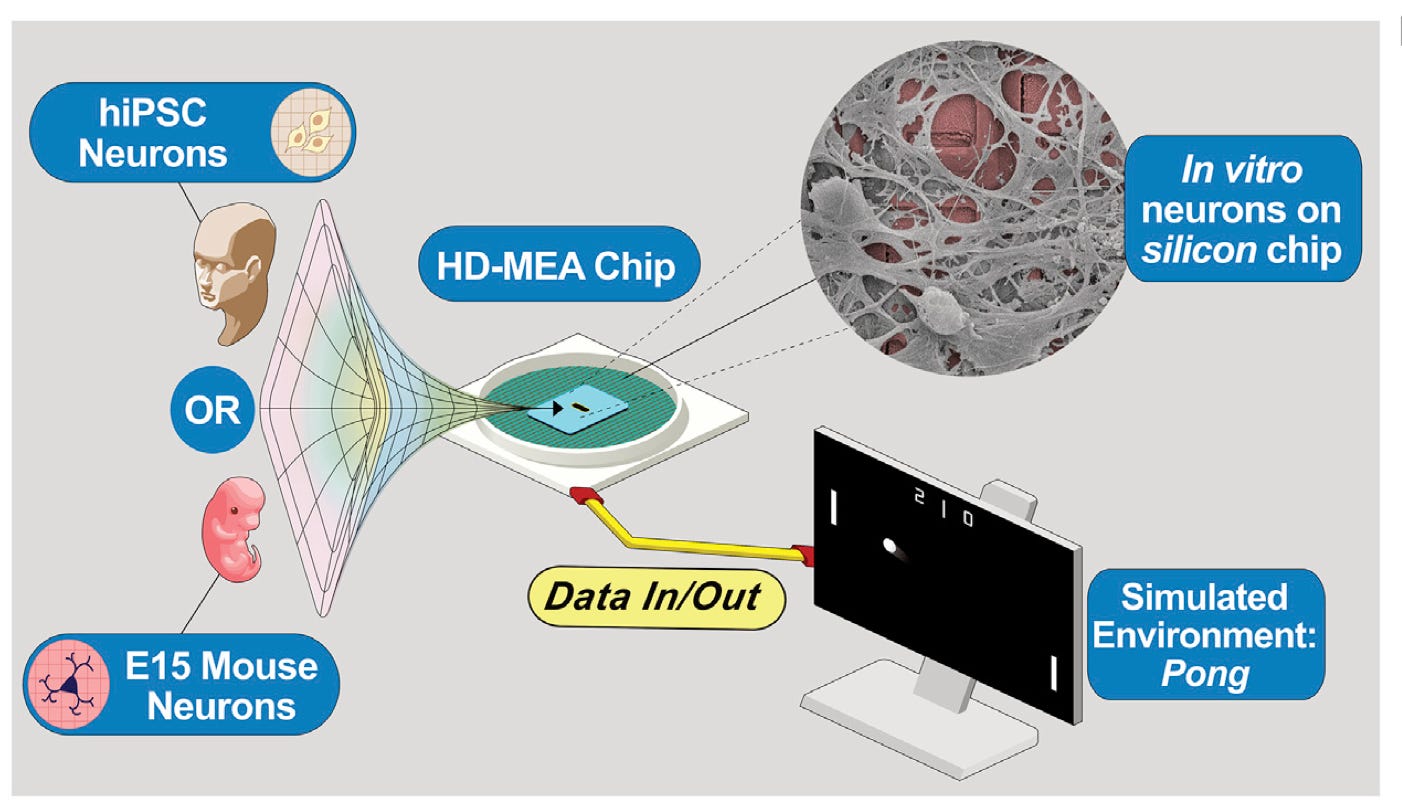
In vitro neurons learn and exhibit sentience when embodied in a simulated game-world [Kagan aet al., Neuron 2022]
Modern deep learning and artificial neural networks, while powerful, are significantly different from biological neural networks (BNNs). There are several advantages of BNNs including energy efficiency, generalization capacity, and network robustness and redundancy. In this paper, Karl Friston’s lab designed a silico-biological hybrid system that learns to play the game Pong, demonstrating the feasibility of training cultured neurons to solve a goal-oriented task. Applications of the approach include testing the effect of drugs on neuronal function and neuromorphic computing platforms with improved computational efficiency and performance. The latest paper from Karl Friston’s (well-renowned theoretical neuroscientist, invented many of the most widely used neuroimaging statistical techniques) lab demonstrates that cultured neurons can learn to play the game Pong. The system is called DishBrain: primary cortical neurons were cultured in a dish, and a multi-electrode array was used to record from and stimulate neurons in a closed-loop system (see schematic below). Sensory information representing the state of the Pong game (e.g., location of the paddle, trajectory of the ball) was communicated to a sensory sub-population of neurons via stimulation using a multi-electrode array. The biological neural network learned to process and integrate this sensory information, and then communicate “action” via a motor sub-population of neurons. If the network’s action resulted in a successful move (i.e., intercepting the ball), a predictable stimulus was delivered to the network; if the move was unsuccessful, an unpredictable stimulus was delivered. This feedback resulted in learning because neurons repeat patterns of activity that result in predictable environments. For more context on this work, and other relevant studies, check out this thread by Lana Sinapayen.
One of the most intriguing (albeit likely far off) applications of this technology is for drug discovery. Enthusiasm for cerebral organoids using induced-pluripotent stem cells in drug development is increasing. Much of the work in the organoid field has focused on recapitulating pathological and molecular changes in neuropsychiatric disease. However, imagine if we could build in vitro models of human cognition? We could test for adverse cognitive side effects of drugs, or develop a disease model for the cognitive symptoms of Schizophrenia. Many hurdles remain, including determining biological plausibility and translatability of these models, but will be a field to watch.
What we listened to
Jack O’Meara, co-founder and CEO of Ochre Bio, sits down with Neil Littman to discuss the large unmet health need created by chronic liver disease and the company’s efforts to develop RNA therapies address it.
Sean Hunt, CTO of Solugen, talks about how Solugen was forced to focus on generating revenue from day one, something extremely difficult for deep tech companies to initially prioritize. He also dives into the evolution of synthetic biology and biomanufacturing and how he had to do as much business model innovation as technical innovation to get Solugen to where it is today.
Hear from a number of leaders in cell therapy to discuss the tremendous advancement in the space, and where the future is heading.
Jack Scannell, the mind behind the term “Eroom’s law”, breaks down methods in drug discovery and how we can make better choices for quality and quantity.
Notable Deals
Illumina joins with AstraZeneca on AI-driven drug discovery project. Illumina’s evolution from tools company to drug discovery juggernaut contines. The AZ partnership will focus on discovering novel targets by applying AI to multi-omic data to identify genes causally linked to disease. The project will leverage PrimateAI, Illumina’s deep residual network for predicting clinical associations of genetic variants, and SpliceAI, a convolutional neural network for predicting splice variants from non-coding mutations. Illumina's other drug development efforts include a partnership five-year collaboration with Deerfield.
Ginkgo inks $144M deal with Merck to engineer enzymes to improve ingredient manufacturing. The partnership will have Ginkgo develop a strain of biocatalysts that can be used in chemical synthesis for the production of APIs.
Prime Medicine, a preclinical biotech developing novel gene editing therapies for various diseases, filed on Friday with the SEC its S-1 to raise up to $100 million in an initial public offering. However, this is likely a placeholder for a deal we estimate could raise up to $200 million. The company is developing one-time curative genetic therapies using Prime Editing technology, and currently has a pipeline of 18 preclinical programs targeting indications such as sickle cell disease, various liver diseases, non-syndromic hearing loss, neuro-muscular conditions, Duchenne muscular dystrophy, and cystic fibrosis. Prime Medicine has established preclinical proof-of-concept in vivo with long term engraftment of ex vivo Prime Edited human cells in mice in its sickle cell disease program (partnered with Beam Therapeutics).
Matchpoint Therapeutics raises a $70M Series A to push forward covalent molecules. Matchpoint is developing an Advanced Covalent Exploration (ACE) platform that uses machine learning, a robust covalent compound library, and chemoproteomics to develop the next generation of covalent therapeutics. Investors include Vertex, Digitalis, Alexandria, and Atlas.
CRISPR pioneer Feng Zhang is building a new company to solve delivery. Arch Venture Partners, GV and Lux have invested in Aera, which has raised about $200 million. The company is based off these two papers (1, 2)
What we liked on Twitter









Events
Building in Bio Hackathon | Oct 20-24th | SF
Join Pebblebed for an exclusively bio focused hackathon in SF from Oct 21-24. Register by Oct 20. Prizes include 3 months of coworking access and an NVIDIA GPU.
Hummingbird x Fifty Years, Bits in Bio Meet Up | Oct 24th | SF
Bringing together hackers, operators, students, investors, and more working at the intersection of software and biotechnology. Food & drinks are provided. See you there!
SYN BIO 2.0 | October 25-26th | Chicago
KdT Ventures is holding a happy hour on Tuesday, October 25th @ Timmy O’Toole’s, just ahead of the Northwestern Syn Bio 2.0 Event on October 26th at the Evanston Campus. Invite to happy hour + event here. Join us – and bring a sweater!!
American Society of Human Genetics | Oct 25-29 | LA
The ASHG Annual Meeting is the largest human genetics and genomics meeting and exposition in the world. Held in October, it provides a forum for the presentation and discussion of cutting-edge science in all areas of human genetics. Highlights include invited symposia; abstract-driven plenary, platform, and poster sessions; education/trainee workshops; and career opportunities and networking events.
San Francisco Founder-Led Bio Launch Party | Nov 3rd | SF
BioFuture 2022 | Nov 7-9th | NYC
“BioFuture is where relentless therapeutic pioneers, innovators, and investors gather to assess and shape the future of healthcare and digital health. Participate in candid, unfiltered discussions. At this year’s summit, we’ll explore the exciting mashup between rapidly evolving fields including biopharma, digital medicine, big data, AI, healthcare systems, payors, and more. The coming decade will dramatically accelerate the transformation of the healthcare ecosystem. Be part of the discussions that will shape and transform the future of therapeutics.” Use code BF22VIPSPK for 15% off.
Field Trip
The Anatomy of the Data Room, Morgan Cheatham, a member of the Decoding TechBio collective, published everything he knows about data rooms! Check out the piece for folder-by-folder breakdowns of what to include in your data room, and learn from the common mistakes section to drive efficiencies in your fundraising process.

Did we miss anything? Would you like to contribute to Decoding TechBio by writing a guest post? Drop us a note at decodingtechbio@gmail.com or chat with us on Twitter: @ameekapadia @pablolubroth @patricksmalone @morgancheatham @ketanyerneni