BioByte 006
countering the weaponization of genetics, imagenet for spatial bio, power laws in biotech vs tech, the gender gap in biomedical research, business models in bio, and more
Welcome to Decoding TechBio, a writing collective focused on the latest scientific advancements, news, and people building at the intersection of tech x bio. If you’d like to connect or collaborate, please shoot us a note here. Happy decoding!
What’s up fam! Quick update: we are moving our weekly cadence to Tuesday moving forward. Your feedback has been incredibly helpful thus far, so keep it coming (you can get in touch by sending us an email).
We thought we’d kick this week off with a classic XKCD comic. Each time you marvel at the potential of AI, just remember: training modern deep learning algorithms is essentially just a bunch of dot products!
What we read
Blogs
Machine learning-powered drug discovery: Now and Tomorrow [Lin Ning, An Ounce of Biotechnology, 2022]
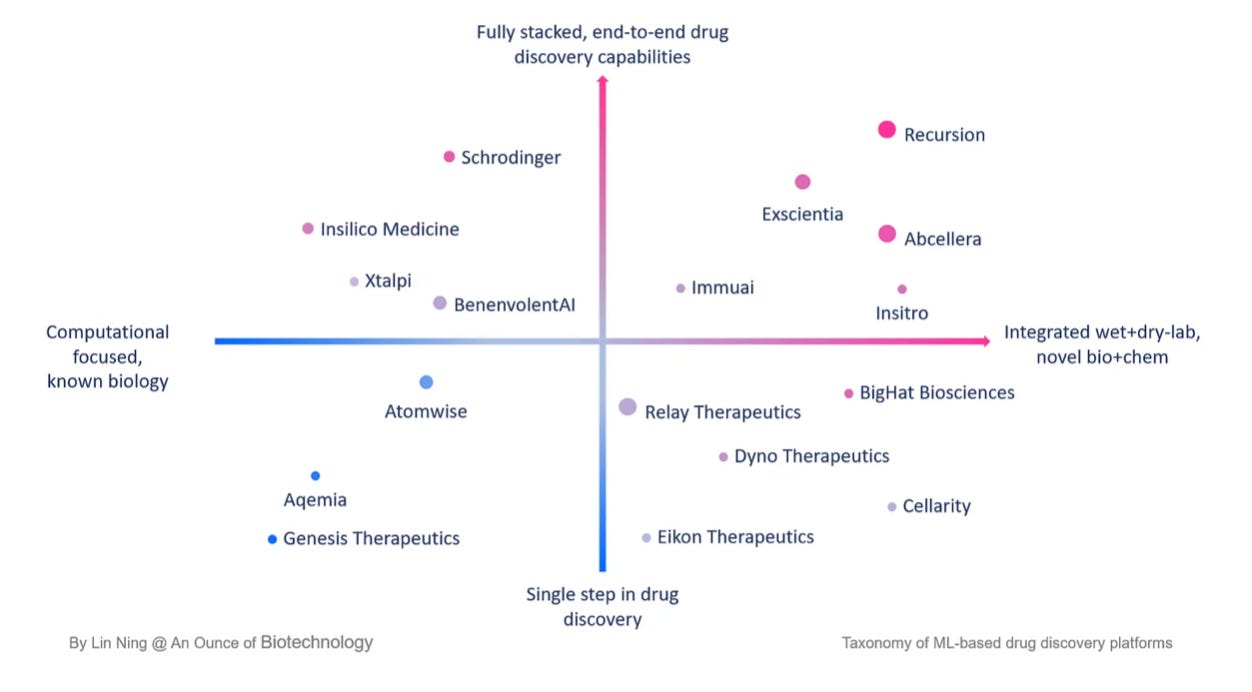
Lin Ning dives into the main characteristics of AI-first drug discovery companies:
Platform-first company structure: for ML-driven drug discovery companies the critical decision isn’t the typical ‘platform vs asset’, but rather how to build a platform that delivers 10x or 100x efficiency. Any AI-first biotech companies who are serious about using ML for drug discovery are enabled by platforms that are composed of high-quality, ML-grade data, wet lab infrastructure, and computing resources.
Versatile business models: it is common to see companies with mixed business models ranging from internal development to including partnerships, spinouts & joint ventures and CRO-type services.
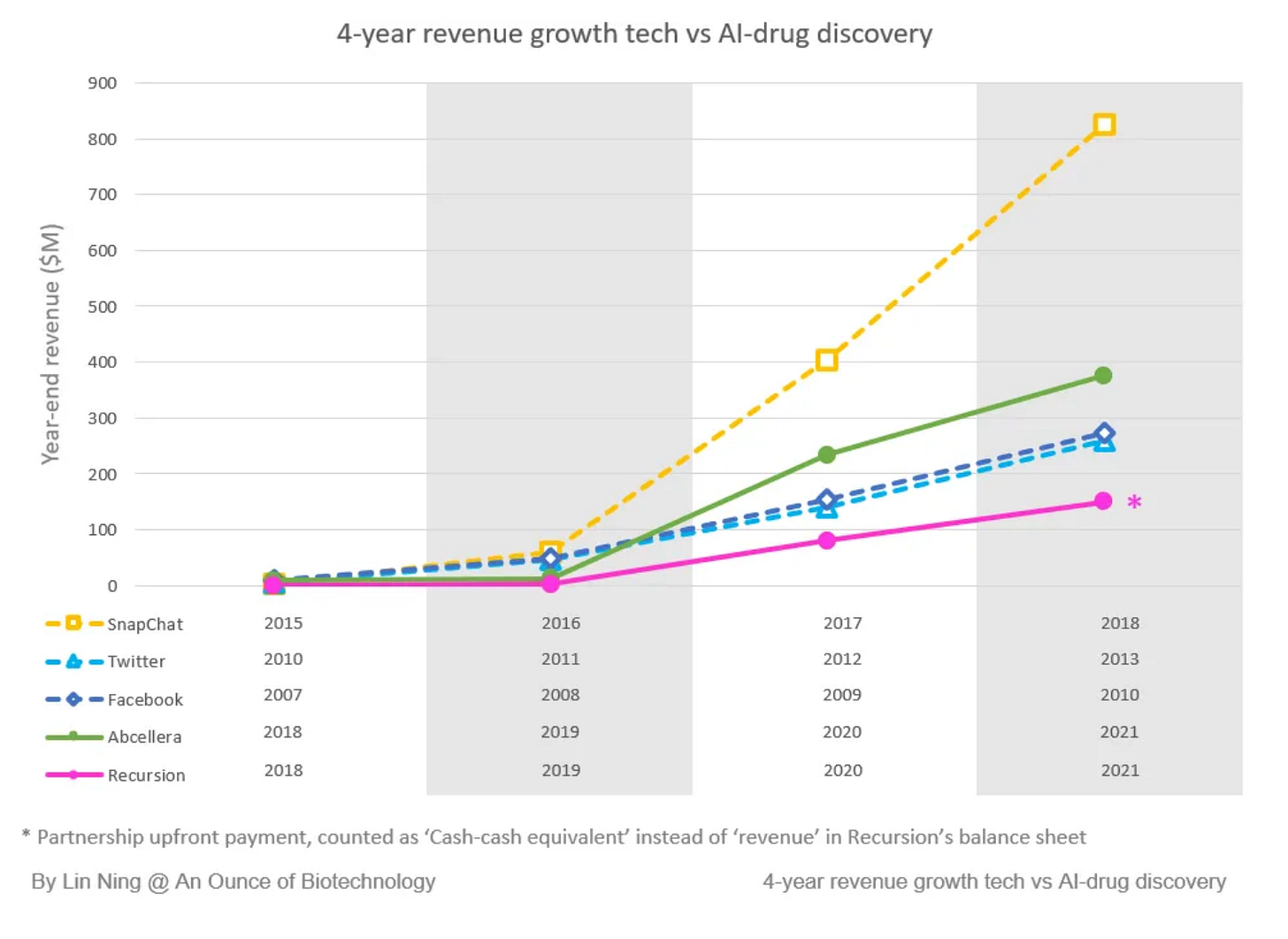
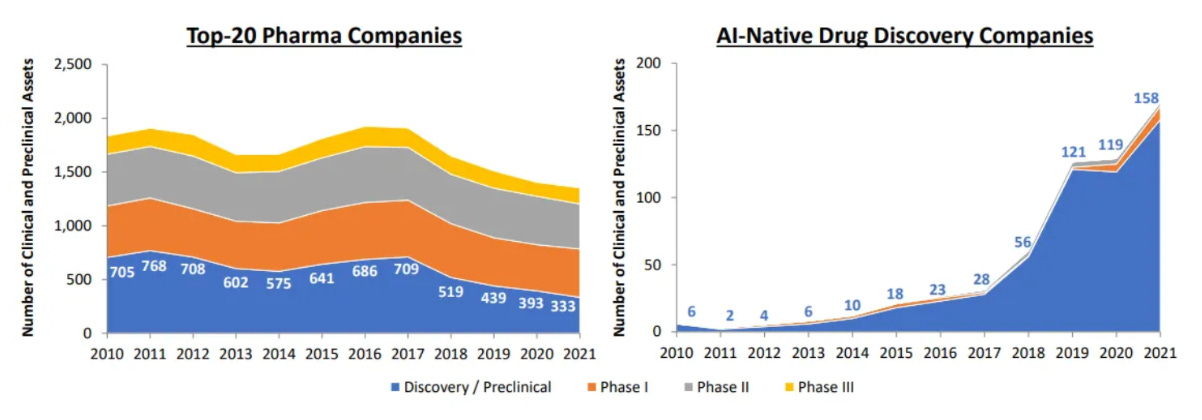
Scaling up like tech companies: By the end of 2021, there were 158 drug candidates in the discovery/preclinical stage and 15 in the clinical stage from the top 20 AI-first companies. In addition, Abcellera’s revenue growth is comparable to Facebook, Twitter, and Snapchat in their early years.
Comparing VC returns in biotech vs. tech [Richard Murphey, Bay Bridge Bio, 2021]
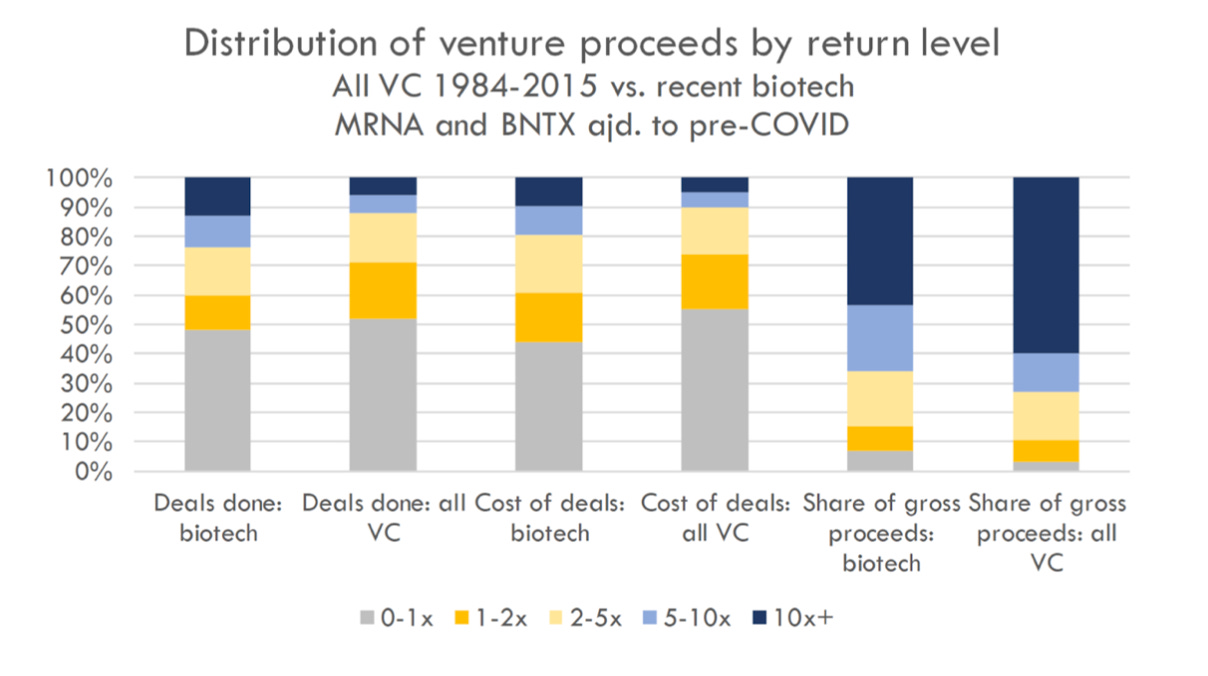
Venture capital is a game of outliers. Venture returns follow a power-law distribution, where the vast majority of profits in a given fund result from just a few investments. The power law is well-known in tech VC, but what about biotech? Does biotech also follow a power law, or are returns more normally distributed? And most interestingly for our community - what about TechBio?
Richard Murphy at Bay Bridge Bio (highly recommended blog) compared biotech venture returns to returns across all sectors to address this question. First, how big are the winners in biotech? Across 7,000 investments from 1984-2014, 6% of venture investments account for 60% of the returns. In recent biotech investments (companies that exited 2018-2021; see post for full methodology), however, 13% of investments account for 43% percent of returns (after adjusting for Moderna, an outlier of outliers, see blog for details). Further, in biotech a greater share of investment profit is driven by 1-5x investments vs. 10x+ investments (compare the 2 bars on the right of the figure below). In other words, in biotech the power law is less pronounced.
Given these data, how should biotech VCs think about portfolio construction to optimize returns? It is possible that we shouldn’t take home-run swings like our tech colleagues. The winners aren’t as big, and singles, doubles, and triples matter more to overall returns.
What about TechBio? Part of the TechBio thesis is that AI-enabled, digital-first platforms scale like software and tech companies. Will the returns for TechBio portfolios look like tech? It may be too early to tell, but anecdotally, early evidence suggests that TechBio companies exit at similar valuations to their non-computationally oriented counterparts. This question is worthy of its own analysis, however, as the answer is likely far more nuanced.
It turns out we need to make (a huge amount of) stuff - Investing in and designing for bio-manufacturing [Bio Endeavors from Innovation Endeavors, 2022]
If as much as 60% of the physical inputs to the global economy are to be produced biologically, we will need to accelerate the scaling and automation of bio-manufacturing processes. And, as therapeutic complexity increases biologically, manufacturing at scale will present key challenges. In this piece, the Innovation Endeavors team offers an overview of biomanufacturing, an analysis of venture investment in synthetic biology by category, and key areas of the biomanufacturing stack — from continuous fermentation to cell-free systems — that show promise for “more economical, flexible, or otherwise better” production.
If you’re interested in this topic, check out President Biden’s National Biotechnology and Biomanufacturing Initiative released in September 2022.
Counter the weaponization of genetics research by extremists [Carlson et al., Nature Comment, 2022]
The co-option of scientific studies by extremist political groups is not new. Over the last few years, meta-research and sociological studies have explored whether human genetics research might be particularly vulnerable. In this fascinating opinion piece, Carlson et al. explore the evidence of the weaponization of human genetics research and propose questions about how human geneticists might respond.
Scientific Publishing: Peer review without gatekeeping [Eisen et al., eLife, 2022]
As of last week, “eLife is changing its editorial process to emphasize public reviews and assessments of preprints by eliminating accept/reject decisions after peer review.” By way of background, eLife is a not-for-profit peer-reviewed open-access scientific journal for the biomedical and life sciences. This announcement is notable for the Open Science movement and calls out flaws in the current state of scientific publishing and peer review. eLife’s alternative system will be governed by three key axioms:
Authors should be able to share their work freely and openly when they think it is ready.
Peer review should consist of scientists publicly sharing their assessments of already published papers, either under the auspices of an editorial organization that oversees the review process, or on their own.
Works of science should be reviewed by multiple relevant groups and individuals throughout their useful lifespan.
Academic papers
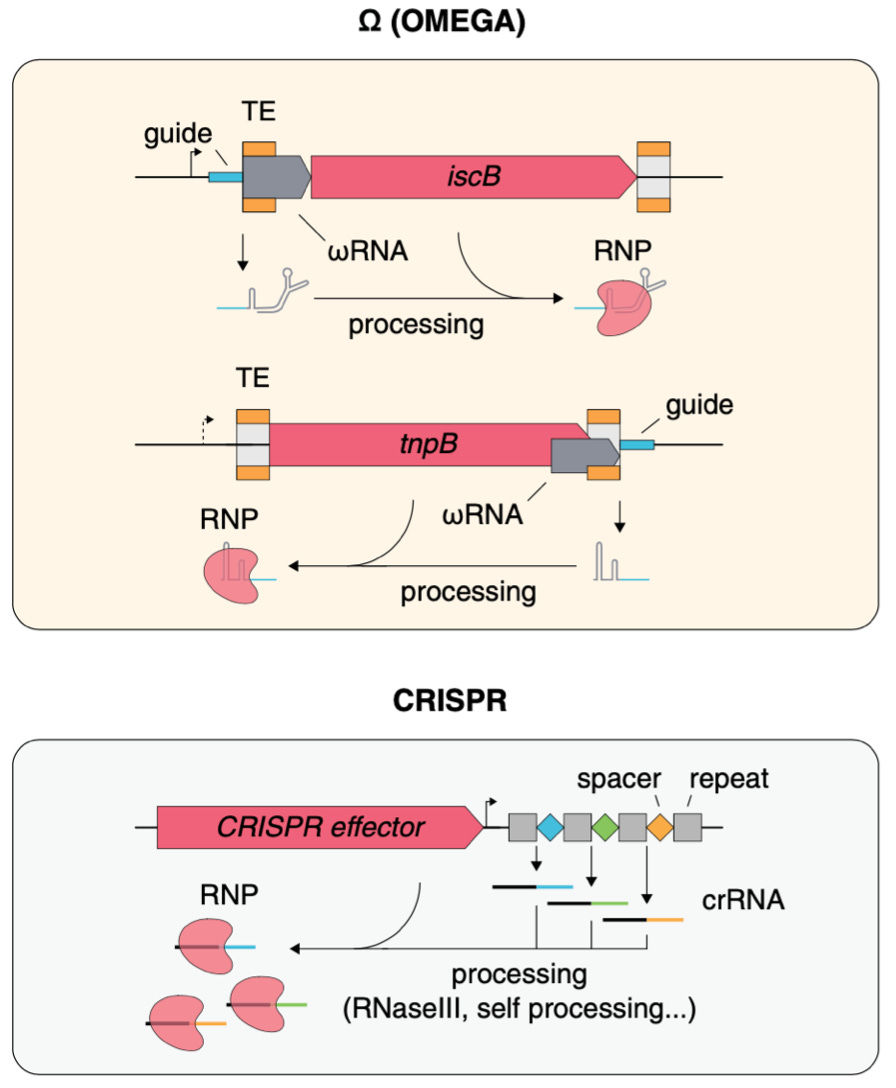
Structure of the OMEGA nickase IsrB in complex with ωRNA and target DNA [Hirano et al., Nature, 2022]
Why it matters: CRISPR-Cas gene editing systems enabled a step change in biomolecular tooling, the treatment of disease and the life science as a whole. OMEGA is a recently discovered widespread class of transposon-encoded RNA-guided nucleases which has distinct properties versus CRISPR-Cas systems. The elucidation of its structure and target-binding mechanism is an important step forward in its adaptation as a tool or therapeutic platform in the future.In 2021, Feng Zhang’s lab identified a new class of RNA-guided systems, termed OMEGA, which include IscB, the likely ancestor of Cas9, and the nickase IsrB, a homologue of IscB lacking the HNH nuclease domain. These enzymes are not part of CRISPR immunity, but the team discovered that each OMEGA system-related protein had a small RNA encoded nearby and it directed the enzymes to cut specific DNA sequences. They named these RNAs ωRNAs.
In this paper, the lab identifies the CryoEM structure of the IsrB complex with its target DNA. Here are some highlights on its structure: IsrB is small in size (350 amino acids) but contains a large RNA guide (300 nucleotides). Instead of using a DNA recognition lobe (REC) like the Cas9 protein, IsrB relies on its ωRNA, part of which forms an intricate ternary structure positioned analogously to REC.
Reliably assessing the electronic structure of cytochrome P450 on today’s classical computers and tomorrow’s quantum computers [Goings et al., PNAS, 2022]
Why it matters: Simulating chemistry is likely to be one the first practical and commercial applications of quantum computers. In this paper, a collaboration between big tech, pharma, and a startup showed what quantum resources would be required to simulate cytochrome P450 enzymes, molecules which are important to drug discovery. A study out last week in PNAS from Google Quantum AI, Boston-based quantum startup QSimulate, and Boehringer Ingelheim identified an application in chemistry with the potential for quantum advantage, or a problem that can be solved by quantum computers but not by classical ones. The search for applications with quantum advantage is a key goal of the quantum computing field.
Cytochrome P450 (CYP450) enzymes are involved in many biological and metabolic processes such as oxidizing drugs, hormones, and fatty acids. Simulating CYP450 enzymes with classical computers is extremely difficult and expensive because the electrons can quickly change spin states based on small changes in the structure of the protein. The physics and math in the paper get pretty complicated, but essentially the authors perform a systematic characterization of the classical and quantum resources required to describe the electronic structure of the CYP450 active site using various methods and algorithms and estimate the runtime for each. Their findings demonstrate the potential for a quantum advantage for quantum-based methods in estimating the electronic structure of CYP450 enzymes.
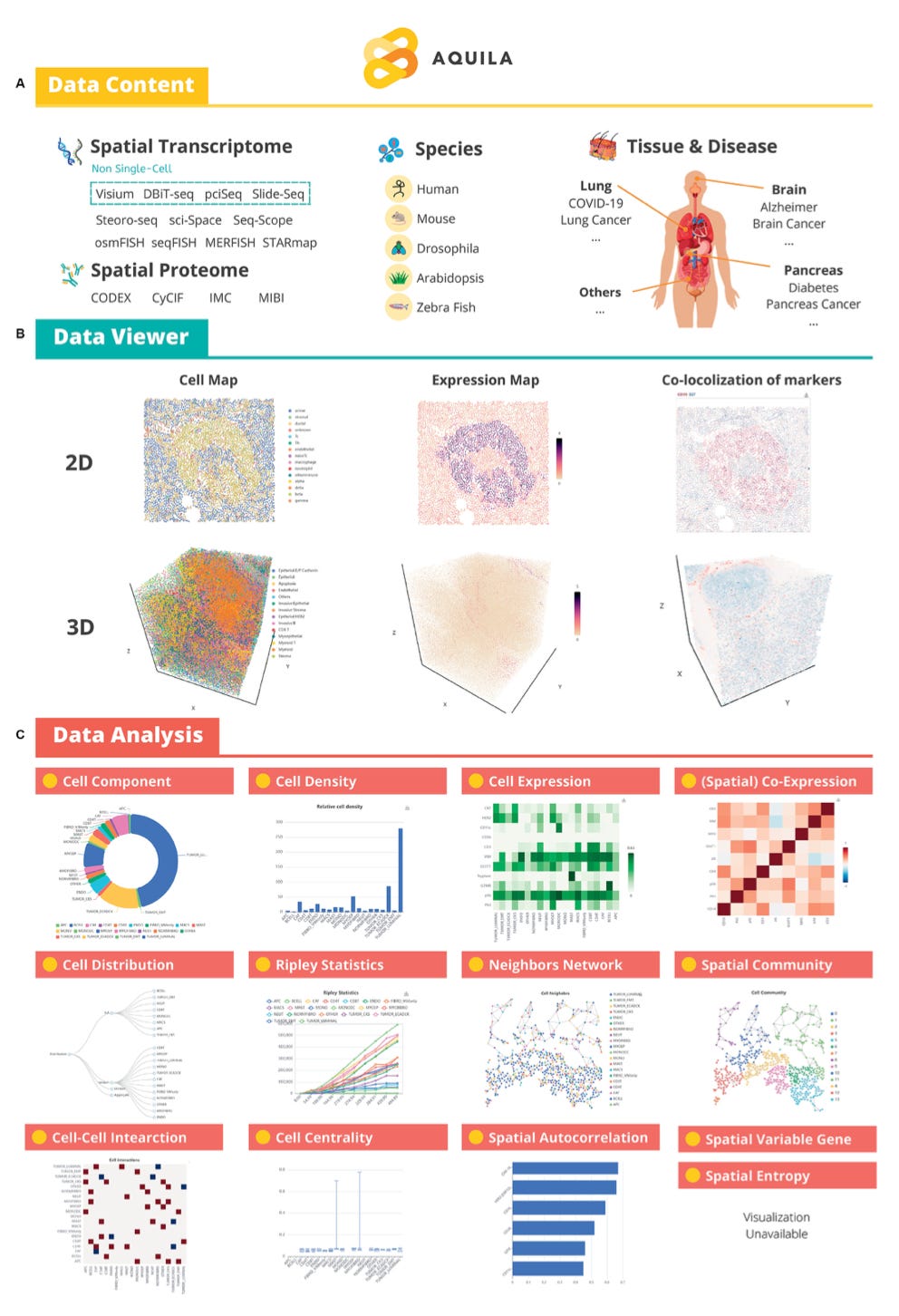
Aquila: a spatial omics database and analysis platform [Zheng et al., Nucleic Acids Research, 2022]
Why it matters: Spatial omics is a rapidly evolving field that leverages computer vision to understand tissue microenvironments and cellular networks. As we saw with ImageNet and the early days of convolutional neural networks, the success of new computational approaches often hinges on a field’s ability to establish robust, open-access data assets for methods development. In this paper, Zheng et al. present Aquila, a spatial omics database and analysis platform featuring 107 datasets from 30 diseases, including 6500+ regions of interest, and 15.7 million cells… AKA ImageNet for Spatial Biology. You can explore Aquila and peruse the tutorial here.
A circulating subset of iNKT cells mediates antitumor and antiviral immunity [Cui et al., Science Immunology, 2022]
Why it matters: The immune system is immensely powerful, and we are still decoding its complexity. Invariant natural killer T-cells (iNKTs) are a type of cell that straddle the bounds of innate and adaptive immunity. Widely considered to be non-circulating and tissue-resident, iNKTs are known to enable tissue-specific protection. The authors identify a new population of circulating peripheral iNKTs that mediate significant antitumor and antiviral immunity.In this paper, Cui et al. used a series of mouse models, transcriptomics, flow, and histopathology to understand invariant natural killer T cell (iNKT) cell population heterogeneity. They identified two unique populations of IL-15-dependent iNKT cells characterized by variable CD244/CXCR6 expression, one of which was peripherally circulating; these cells demonstrated significant cytotoxicity and protection from melanoma metastasis and influenza infection.
No complex machine learning - but rational identification of potent cell subpopulations with carefully planned and executed experiments will continue to feed into future cell therapy efforts.
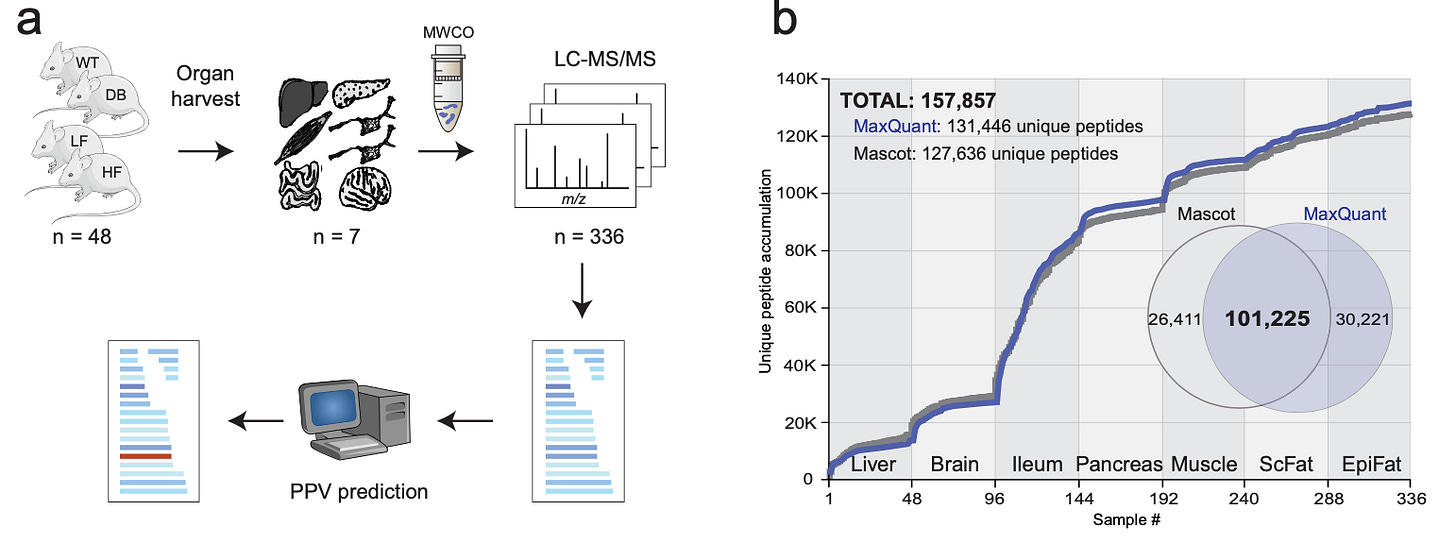
Combining Mass Spectrometry and Machine Learning to Discovery Bioactive Peptides [Madsen et al., Nature Communications, 2022]
Why it matters: Peptides are biologically active molecules that play key roles in biological process regulation, informing the biological underpinning of about 80 marketed drugs (e.g., GLP-1) and 150 clinical development candidates. Though tens of thousands of peptides have been documented in the literature, our current understanding only includes ~300 confirmed bioactive peptides in humans (the rest are largely inactive degradation products). Mapping and expanding the known, biologically active peptide space using high-throughput omics technologies and mass spectrometry is an exciting area for drug discovery – after all, the first peptide drug was insulin!In this paper, Madsen et al. conducted a large-scale peptidomics analysis of seven different organs in four mouse strains. Machine learning algorithms applied to the peptide structures facilitated clustering to predict the most likely endogenously functional peptides, separating them from those that were inactive degradation products. Initially, the algorithms demonstrated an ability to identify many of the already-known bioactive peptides. Leveraging prediction-based methods with amino acid sequences observed from mass spectrometry data, the team then identified several new candidate peptides and validated the bioactivity via in-vitro and in-vivo screening.
The Gender Gap Amongst Doctoral Students in the Biomedical Sciences [Schaller, bioRxiv, 2022]
Why it matters: This critical study explores gender inequity in the biomedical sciences with a focus on the disparity in research productivity between men and women across 42,000 doctoral students working with over 16,000 advisors at 235 institutions in the United States. Researchers identified the following disparities in research productivity:
Men produce >10% more first-author papers
Men produce >15% more total papers
First-author papers produced by men receive >17% more citations
Offering numerical insight into gender-based disparities in biomedical research, these data when published will be well-positioned to support new initiatives that support women and non-binary scientists at the formative stages of their careers.
What we listened to
Image-Activated Cell Sorting with Keisuke Goda [Axial Podcast]
A fascinating interview with Keisuke Goda, a physicist-turned-biologist from the University of Tokyo that is a pioneer in image-activated cell sorting, which combines flow cytometry, microscopy, and machine learning, to do cell sorting in real-time and at high throughput. See also Josh Elkington’s blog on the method.
Oncology Discovery Advances with AbbVie’s Steve Davidsen, Ph.D., [Business of Biotech]
This podcast interview features the story and perspectives of Dr. Steve Davidsen, Vice President of Oncology Discovery at Abbvie, where he joined in 1986! Steve offers an overview of 36 years in oncology drug discovery and how he’s evolved the company’s research strategies by implementing advanced technologies over the years, and why he’s now focusing on computational biology.
Human brain organoids implanted into rats could offer a new way to model disease, [Thompson, Nature Podcast]
In this podcast, researchers discuss “neural organoids'' and the role they have played in researching brain development and disease progression since the early 2000’s. Challenged by limitations due to structural differences between these organoids and the complex hierarchy of the human brain, researchers are now transplanting these organoids into the brains of existing animal models.
For more on the approach of transplantation of human cortical organoids, check out this new paper by Revah et al. in Nature.
Notable Deals
GSK expands AI partnership with Tempus, fronting $70M for access to multimodal data. The companies announced a three-year extension of their deal which started in 2020. The new deal will focus on “improving clinical trial design, speeding up enrollment and identifying drug targets.” In order to do this, the AI team at GSK will leverage Tempus’ dataset; which includes “de-identified patient data, which provides clinical information such as the tumor sequence, pathology slides, imaging aspects, and circulating tumor DNA. On top of that, the dataset includes clinical outcomes.” You can have a look at our analysis on multimodal healthcare data in our past TBWIR edition here.
Healthcare MLops and federated learning platform Rhino Health announces $6.7M Seed extension. Rhino Health is a federated learning platform that allows AI developers and medical researchers to access diverse datasets in a privacy-preserving manner. The geographic distribution of data used to train clinical AI algorithms is remarkably homogenous; of a sample of published healthcare AI papers, 34 states in the USA were unrepresented in the training data, potentially introducing a risk of bias. Rhino Health’s platform manages the full life cycle of healthcare AI, from aggregating training data, to model development, deployment, and monitoring.
What we liked on Twitter







Events
Jefferies London Healthcare Conference | Nov 15-17th | London
“This year marks the 13th anniversary of the largest healthcare-dedicated conference in Europe. In 2021, we hosted over 650 healthcare companies, 3,000 attendees, and nearly 8,000 investor and business-to-business meetings. Our conference will again feature leading public and private companies from the pharmaceuticals, biotechnology, medical technology, and healthcare services sectors from the Americas, Europe, Middle East, Africa, Asia, and Australia.” List of participating companies here. You can email the organizers here if you’d like to get an invite.
Field Trip






Did we miss anything? Would you like to contribute to Decoding TechBio by writing a guest post? Drop us a note here or chat with us on Twitter: @ameekapadia @ketanyerneni @morgancheatham @pablolubroth @patricksmalone