BioByte 010
advances in neuromorphic computing, a large language model for science, FDA innovation in cell based meat, improvements in design of clinical trials, intratumoral microbiota in cancer and more
Welcome to Decoding TechBio, a writing collective focused on the latest scientific advancements, news, and people building at the intersection of tech x bio. If you’d like to connect or collaborate, please shoot us a note here. Happy decoding!
Happy Wednesday! Hope you enjoy this densely packed week in review ahead of Thanksgiving. Best served with gravy.
We have some long reads cooking in the oven that we’re excited to release in the coming weeks — stay tuned! Want to write something with us? Email us here.
What we read
Blogs
New Chip Expands the Possibilities for AI [Allison Whitten, Quanta Magazine, 2022]
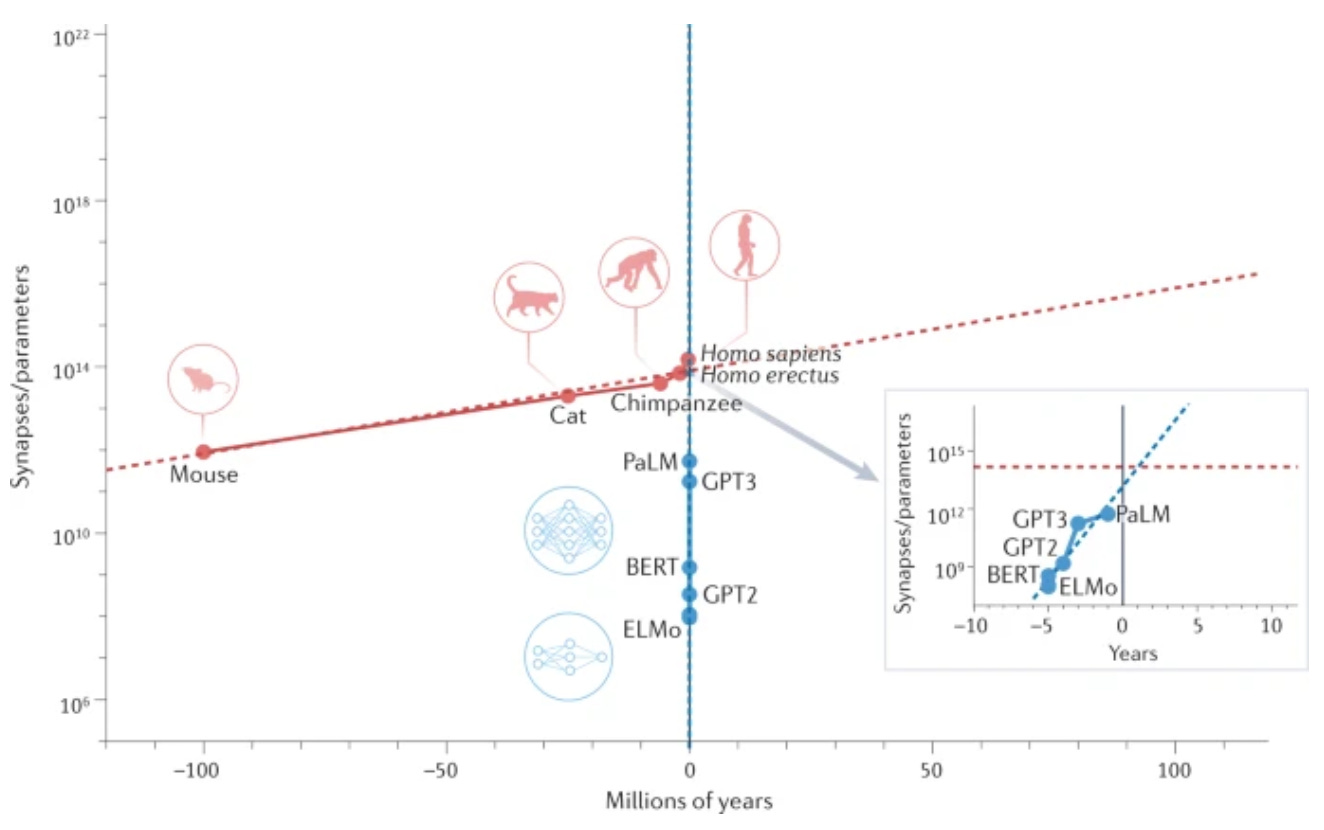
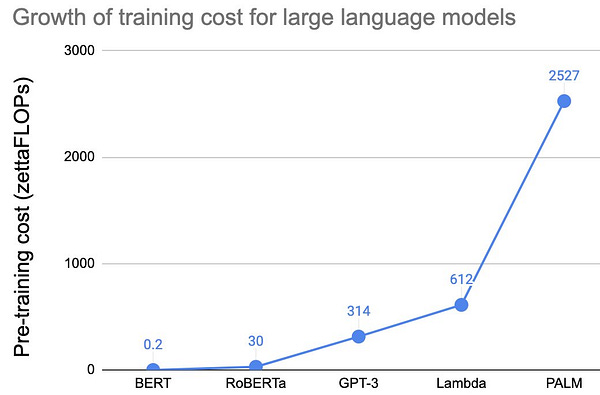
Why it matters: The cost (both economic and for the environment) of modern AI is significant and unsustainable. A recent breakthrough published in Nature details a new neuromorphic computing chip that performs as well as traditional hardware on complex AI tasks, while being 1000x more energy efficient.Modern AI, especially deep learning algorithms, are incredibly expensive and resource intensive to build and train. The training of a transformer-based algorithm emits 5x the amount of carbon dioxide emitted by an average american car. The estimated cost of training Open AI’s GPT-3 is about $4.6M, with estimates of newer large language models pushing north of $20M. These costs are not sustainable.

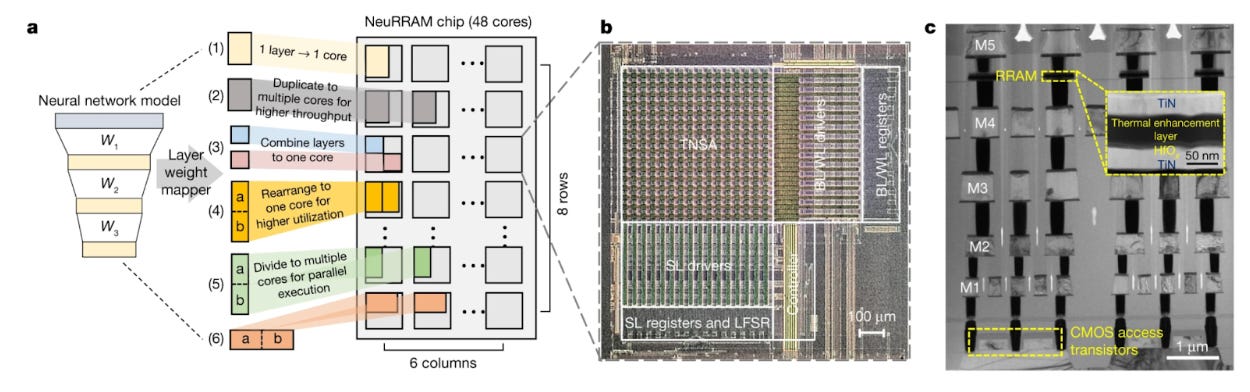
Researchers have long looked to neuromorphic computing (NMC) as a potential workaround to energy-inefficient hardware like the traditional Von Neuman architecture or GPUs. NMC is a hardware architecture modeled after energy-efficient electrical circuits present in the nervous system. Despite its promise, computational performance of NMC, especially on AI tasks, has significantly lagged modern deep learning running on traditional digital computers.
Weier Wan and colleagues published the details in Nature of a new neuromorphic chip called NeuRRAM. NeuRRAM uses a new type of analog-based memory called resistive RAM. Unlike typical binary (1 or 0) RAM, resistive RAM stores values along a continuum and therefore can store more information. Further, rather than transferring these values from RAM to a central processing unit, basic compute tasks take place at the site of storage. The result is a chip performs as well as traditional hardware on complex AI tasks, while being 1,000 times more energy efficient.
Technical challenges remain, however, before widespread adoption. There is an element of randomness in analog computing that introduces noise and makes it more difficult to run deterministic AI algorithms. The NeurRRAM chip can model millions of weights, short of the billions of weights required to run a large language model like GPT-3. Future work will focus on scaling up this architecture.
Atoms are local [Elliot Hershberg, The Century of Biology, 2022]
Let yourself dream about the possibilities of biology and building with atoms with Elliot’s piece on the industrialization of biotech. He ventures down a few thought experiments to paint a clear picture of what the future could look like when you architect with biology. For example, if fungi can cover entire spans of land, why wouldn’t we be able to grow a treehouse with biology? Elliot’s article is a great reminder that industrial biology and the bioeconomy aren’t siloed to fermentation and bioreactor driven processes. Sure making industrial compounds cheaper and greener is a massive step forward in bioinnovation, but it nowhere near scratches the surface of what is possible with biological building blocks. Elliot closes by reiterating that atoms are local. This simple truth underscores how scale and distributed manufacturing of biology is possible.
Founder-led biotech is not a new concept. Impressive companies emerged this way [Alex Schubert, 2022]
Founder-led, as opposed to hired-management led, biotech companies have emerged over the years to create unlikely, yet incredible outcomes. The founders were met with skepticism from investors but persevered nonetheless. Alex writes about some of the greatest hits in this piece: Gilead, Regeneron, Vertex, Genentech, BioNTech, Kite Pharma, 10x Genomics and Ginkgo Bioworks.
FDA Spurs Innovation for Human Food from Animal Cell Culture Technology [FDA, 2022]
This past week, the FDA announced completion of the first pre-market consultation of a human food made from cultured animal cells submitted by UPSIDE Foods, and reported no further questions about the organization’s safety conclusion. There’s still a ways to go before UPSIDE’s products hit the shelves including additional review of the manufacturing facilities and processes by both the U.S. Department of Agriculture and FDA. This news is exciting for food developers leveraging cell culture technology with animal cells obtained from livestock, poultry, and seafood.
Academic papers
Should artificial intelligence be interpretable to humans? [Matthew Schwartz, Nature Rev Physics, 2022]
Why it matters: Artificial intelligence algorithms are increasingly playing the role of scientist, but their discoveries aren’t always human-comprehensible. Should they be? What happens when scientific discoveries advance to the point that they are no longer understandable by humans? It is commonly expected that scientific theories should be comprehensible by humans. This is an understandable assumption - science is the process by which explanations are generated about the natural world. What good are explanations if we as humans cannot understand them?
Artificial intelligence, and large language models (LLMs) in particular, are increasingly playing the role of scientists. For example, Google’s PaLM LLM and Minerva Project fine-tuned PaLM to solve quantitative problems in biology, chemistry, and physics. The algorithm was also (partially) interpretable and could explain its answers with text and equations. This raises several important questions: what role will AI play in the scientific method? Does scientific knowledge discovered by AI need to be human comprehensible?
Schwartz pushes back on the expectation that AI should be interpretable. Statistical algorithms have different ways of understanding data than we do. As the complexity of AI models continues to grow, humans are no more likely to understand their inner workings than a cat is to understand calculus. Schwartz argues that letting go of our desire for understanding is similar to other discoveries in the history of science that have forced us to confront our insignificance, such as the realization that the earth is not the center of the universe, or that this dimension may be one of many parallel universes.
Schwartz’s comment is a response to a Perspective published by Mario Krenn et al., in which the authors outline a framework for how AI can contribute to the acquisition of new scientific understanding. A key feature of this framework is the transfer of knowledge, or understanding, between AI and human scientists. My own perspective is that the role of interpretable AI in science depends on the goal: prediction vs explanation. The most accurate predictive models are often the least interpretable (this is related to the bias-variance tradeoff in machine learning), and vice versa. If the goal is to leverage AI to contribute to scientific knowledge, then human-comprehensible algorithms are critical. If the goal is to build a predictive model where accuracy is paramount, interpretability matters less.
Check out the thread below and Ted Chiang’s short story if you are interested in the role of AI in the scientific method.
Improving combination drug trials using ‘definitive screening designs’ [Dodds et al., Nature Biotech, 2022]
Why it matters: the current approach to clinical trial design is constructed for the development of single-agent therapies and it lacks scalability needed to explore the entire space of combination therapies. Full factorial designs that explore all combinations and dosages would be impossible given the resource constraints. The authors give an example on why changing trial design is important:
Consider a hypothetical clinical trial combining n = 4 possible drugs (A, B, C and D), each with d = 3 possible dose levels (no dose, half of the labeled dose and a full label dose). A full factorial design would require 81 (d^n = 34) treatment arms.
Designed experiments can reveal the effects of each cocktail component (called ‘main effects’) and the relatively few combinations that yield synergistic leaps in efficacy (called ‘interaction effects’). The authors explain how ‘definitive screening designs’ (DSDs), a type of experiment design, could reduce the number of treatment arms:
[…] requires just 9 (2n + 1 = 2 × 4 + 1) treatment arms […] DSD designs are very efficient in detecting the primary effect of each treatment (“does drug A work alone?”) as well as treat- ment–treatment interactions (“do drugs A and B synergize?”) without confounding. More complex interactions (“does drug C only serve to potentiate the effects of an A + B synergy?”) may be confounded, but these limitations are known at the time of study design and alternative methods could be weighed.
This methodology, however, still comes with its limitations such as not accounting for patient biology heterogeneity. We do expect that with better combination trial design, unexplored combinations will lead to unexpected cocktail therapies which have brought so much relief to patients for the management of HIV/AIDS, chemotherapy and hepatitis C.
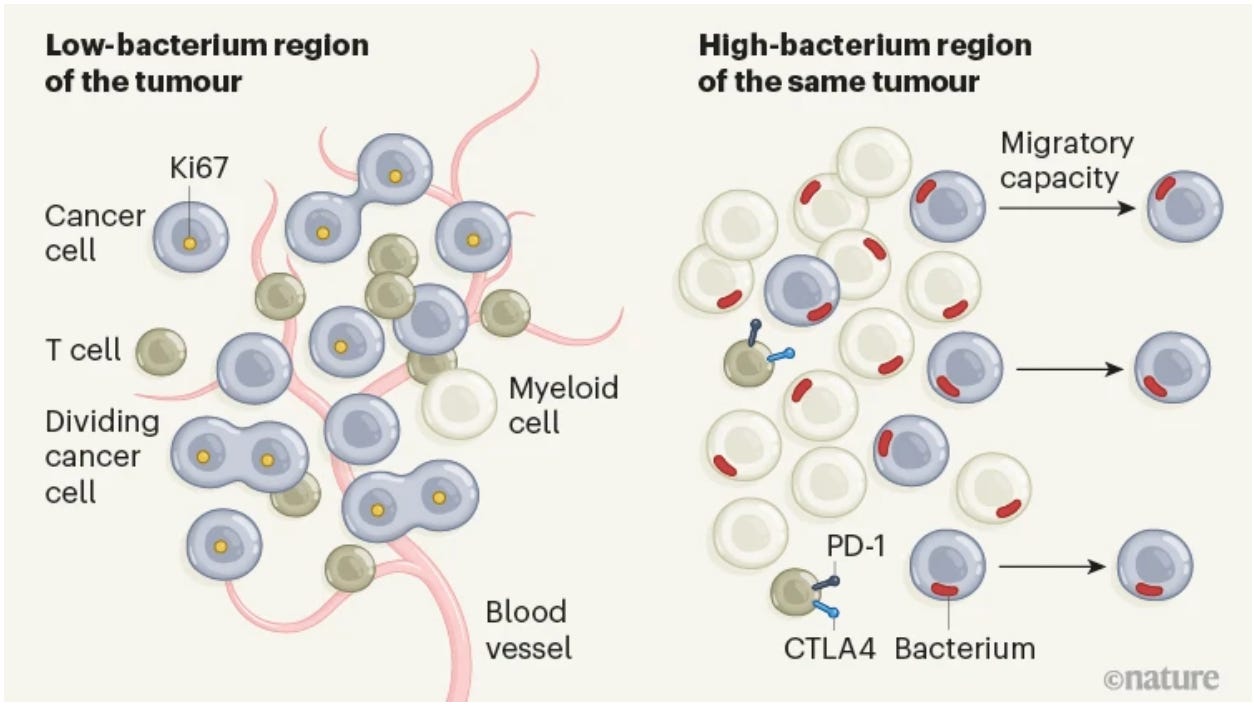
Effect of the intratumoral microbiota on spatial and cellular heterogeneity in cancer [Niño et al., Nature, 2022]
Why it matters: We’ve known for decades that cancer is an incredibly heterogeneous and complex entity, with bacteria, fungi, and viruses playing a role – the question is, to what degree? Here, Niño et al., reveal that tumor-associated microbes are organized in microniches that interplay with immune and epithelial cell states to promote cancer progression.The tumor microenvironment is an ecosystem that is tightly maintained by blood vessels, immune cells, and microbes. For years, the presence and role of bacteria in tumors has been increasingly revealed to be complex and important for disease progression. Here, the authors use RNAscope, which allows visualization of RNA molecules in individual cells, across oral squamous cell carcinoma and colorectal cancer samples. Subsequently, spatial proteomics were identified using microscopy analyses + GeoMX, while spatial transcriptomics were studied using 10X Visium. However, these don’t provide single-cell resolution; thus, the authors developed an RNA-sequencing technique called INVADEseq, which allows human transcripts to be sequenced in parallel alongside bacterial rDNA (ribosomal DNA) transcripts. Long story short, the authors found that the presence or absence of bacteria correlated with disease characteristics: for example, tumor areas containing bacteria were more immunosuppressed (with key proteins, such as PD-1 and CTLA-4 being highly expressed). Interestingly, bacteria were more common in tumor cells that had an abnormal number of chromosomes than in those with normal chromosome compositions. This work plugs in beautifully to the concept of truly personalized medicine, where perhaps patient tumors can be assessed for the classic markers of immunosuppression, but also in bacterial composition that may prognosticate response to therapy.
What we listened to
Shehnaaz Suliman, CEO of ReCode Therapeutics, on lipid nanoparticle delivery of genetic medicines.
Keith Flaherty is the Director of Clinical Research at Mass General Cancer Center and a Professor of Medicine at Harvard Medical School. Dr. Flaherty is a medical oncologist and has worked in the field for 22 years, during which he has founded seven biotech companies. He joins Rahul for an in-depth conversation about his work in oncology. They cover a wide range of topics including his early decision in school to switch from neuroscience to oncology, his entrepreneurial journey and what he’s learned along the way, his perspective on the immuno-oncology landscape and opportunities that lie ahead in this field, what he values in a board and how you can be most effective as a board member, and much more.
Notable Deals
Protein programmers get a helping hand from Cradle’s generative AI. Cradle Bio raised a $5.4M seed round from Index Ventures, Kindred Capital, and angel investors including John Zimmer, co-founder and president of Lyft, and Emily Leproust, CEO of Twist Bioscience. Cradle uses generative AI to reverse engineer proteins with desired structure and properties such as thermostability, binding affinity and specificity, and more.
Simulations for experiments: Bayer-backed startup lands $20M to test out its tech. Turbine raised its Series A round led by Mercia and MSD Global Health Innovation, an investing arm of the American Merck, and joined by Day One Capital and existing investors Accel, Delin Ventures and XTX Ventures. “Turbine’s system consists of what Nagy calls a wiring diagram, or a mathematical representation of protein-protein signaling, based on parameters of protein interactions as decoded by machine learning. Then its engineers take data that have been generated from real cells — genomic and transcriptomic data from liquid biopsies, tumor models, cell lines and so on — and feed them into the program, so that when users run simulations on the platform, they would get essentially the same results as a physical experiment.”
Exscientia, MD Anderson Collaborate on AI-Driven Oncology Drug Discovery and Development. The researchers at MD Anderson’s Therapeutics Discovery division will use Exscientia’s expertise in building machine learning models to help identify druggable targets and in what it terms a “data-agnostic” approach to drug design. As potential therapies are identified in this partnership, the two parties may advance these compounds into proof-of-concept clinical trials at MD Anderson.
Ionis partners up with Metagenomi on drug discovery pact, shelling out $80M upfront. The “first wave” of the partnership is primarily focused on gene editing for metabolic disorders in the liver. The “second wave” will be focused on organs outside the liver: “we’re thinking about everything from kidney to lung to muscle to CNS.”
Insilico Signs Six-Drug AI Discovery Deal With Sanofi Worth Up To $1.2 Billion. Insilico will receive up to $21.5 million in upfront and target nomination fees for each of the six targets. It will also be eligible for milestone and royalty payments. Insilico will apply its end-to-end Pharma.AI platform to the Sanofi targets, including interdisciplinary drug discovery scientists who will advance lead therapeutic compounds up to candidate development stage.
Following secretive AbCellera pact, Moderna picks up another antibody discovery deal via Harbour BioMed. Harbour BioMed will use its antibody discovery platform to identify nucleic acid-based oncology immunotherapies for Moderna, an RNA company. Moderna will own an exclusive sub-licensable license to exploit a panel of sequences against multiple targets, derived from HBM’s proprietary heavy chain only antibody discovery platform. Moderna will be responsible for all development costs, while Nona Biosciences, an HBM subsidiary, will receive upfront and milestone payments plus royalties.
What we liked on Twitter










Events
OTAT Town Hall: Cell Therapy Chemistry, Manufacturing, and Controls | Virtual | December 7 12:00-1:30pm ET]
The FDA’s Center for Biologics Evaluation and Research (CBER) Office of Tissues and Advanced Therapies (OTAT) is hosting a virtual town hall to answer stakeholder questions related to cell therapy chemistry, manufacturing, and controls (CMC), including tissue-engineered medical products regulated by OTAT. This event is part of a series to answer questions from stakeholders about a variety of topics on which OTAT has regulatory oversight.
Field Trip
Gentleman puts out some of the best deep house mixes on Youtube.
The Bicycle Bank Robber. One of the less common paths out there, cyclist → bank robber. Take a dive into the life of Tom Justice, cyclist and bank robber extraordinaire.
The biggest company in the world [Vijay Pande & Daisy Wolf, a16z Bio, 2022]
A16z ascertains that the biggest company in the world will be a consumer healthcare company. The math is obvious: the US healthcare market = 5x global advertising industry. The latter makes most of the revenue for Google, Meta & Co. Current large healthcare companies, like UnitedHealth Group have abominable NPS, capping its absolute size potential. The fund makes two predictions about how a company could get there:
Building a Payvidor with sleek customer experience: the integration of a payer and provider with an Apple-like focus on consumer, would undoubtedly be the preference for anyone searching for insurance.
Building an infrastructure layer: in the US, if you’re looking for a service the search and aggregation leaves a lot to be desired. That’s why the Amazon for healthcare could bring a better experience by allowing consumers to browse and pick from all available options for healthcare services, treatments and professionals. The Visa of healthcare would radically improve healthcare payments to bring an industry that works through fax (!) to the 21st century. In fact, several companies like Zocdoc and Solv have taken notice and have made it easier to shop and pay for services.
This is not a TechBio company, but we think efficiency improvements across the whole value chain will improve the drug discovery process through, for example, better data collection and structuring, leading to a lower attrition rate in drug discovery through better patient stratification.
Did we miss anything? Would you like to contribute to Decoding TechBio by writing a guest post? Drop us a note here or chat with us on Twitter: @ameekapadia @pablolubroth @patricksmalone @morgancheatham @ketanyerneni