BioByte 011
de-noising diffusion models, tumor aneuploidy for prognosis, APOE4's impact on cholesterol in Alzheimer's, and nuclear magnetic resonance imaging for single amino acid residue data
Welcome to Decoding TechBio, a writing collective focused on the latest scientific advancements, news, and people building at the intersection of tech x bio. If you’d like to connect or collaborate, please shoot us a note here. Happy decoding!
Hello from NeurIPS! A few of us are hanging out in New Orleans this week. We had a blast at the Machine Learning for Health event yesterday and will be poster surfing at NeurIPS today and tomorrow. Drop us a note if you’re in New Orleans and want to chat comp bio and meet up IRL!
What we read
Blogs
Denoising Diffusion Generative Models in Graph ML [Michael Galkin, Towards Data Science, 2022]
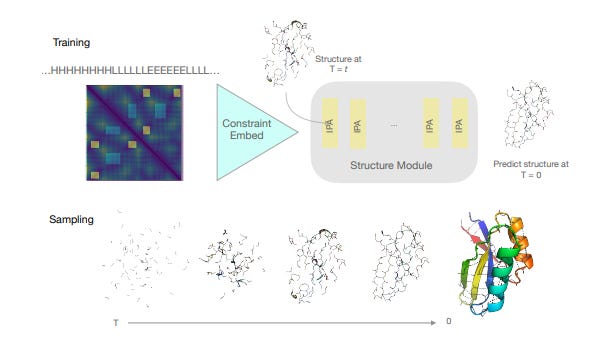
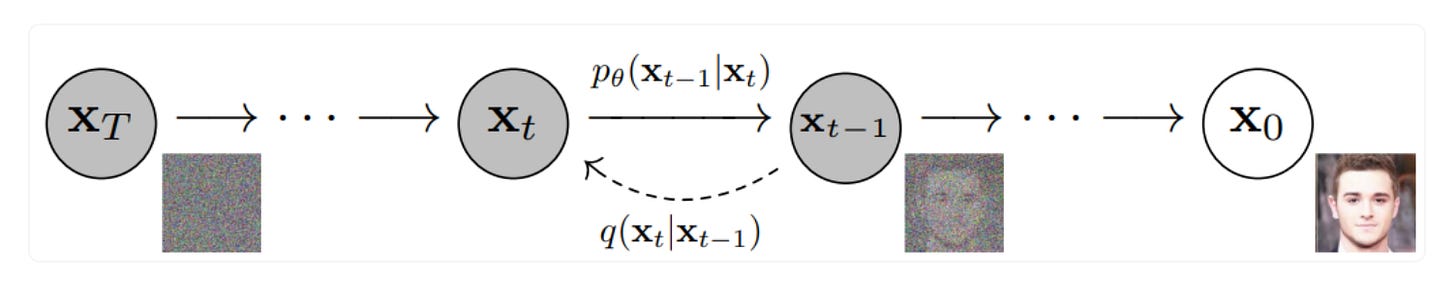
The pace of progress in machine learning in recent years has been WILD. You have no doubt seen the latest, awe-inspiring demonstrations of generative AI systems, such as DALL-E-2, Stable Diffusion, or Imagen. Much of the excitement in generative AI is due to a breakthrough method called Denoising Diffusion Probabilistic Models. So-called diffusion models have quickly surpassed generative adversarial networks (GANs) as the workhorse generative model, especially for image generation. Briefly, diffusion models function by iteratively adding gaussian noise to training data (top figure), and then learning to recover (or generate) the data by reversing this process (bottom figure).
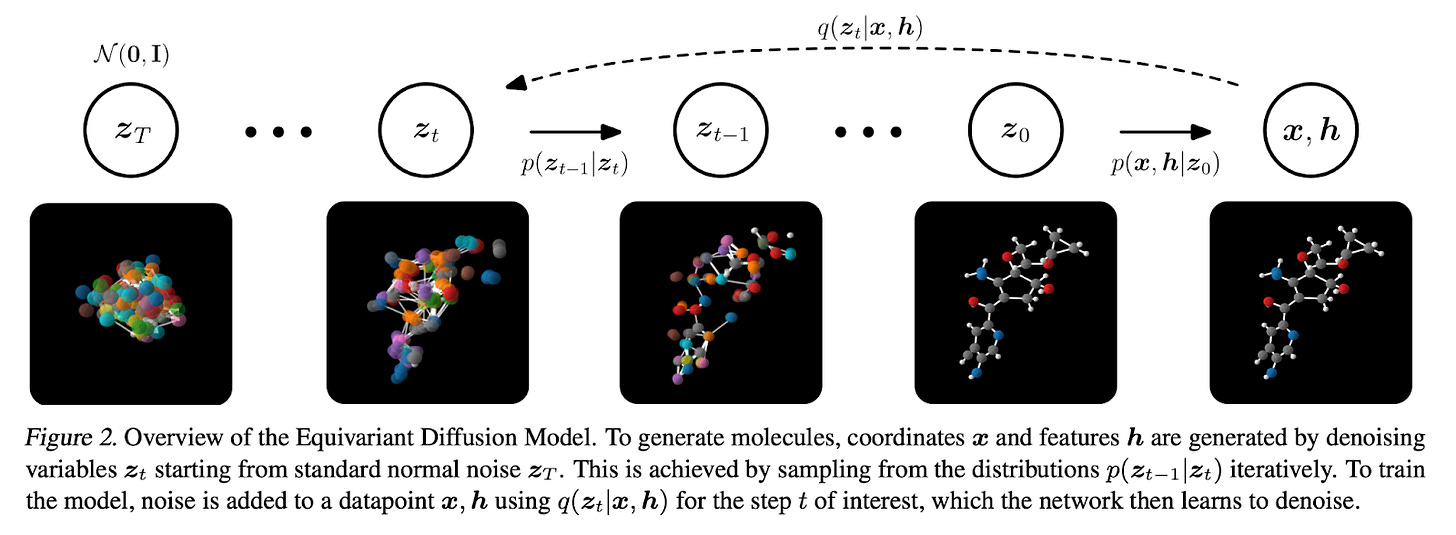
After the diffusion model is trained, new data is generated by passing randomly sampled noise through the trained model. For the mathematically curious, check out this explainer for more detail. In addition to images and text, diffusion images can be used to generate molecules. Earlier this year, Emiel Hoogeboom and colleagues designed a diffusion model that outperformed previous 3D molecule generative methods in both quality of generated molecules and computational efficiency of model training. The input to the diffusion model was a 3D graph representing the chemical structure of the molecule. Nodes of the graph represented the 3D coordinates and types of atoms, while edges between nodes represented bond types. In the forward diffusion process (right -> left in the figure below), noise is injected into the graph of the molecule until we have a random ball of atoms. Then, in a reverse diffusion process (left -> right in figure), the model learns to predict and subtract the noise to generate the original molecular graph. Finally, once these distributions are learned, the method can be extended to generate molecules with desired structure and properties such as optimal polarizability and heat capacity.
In the blog post, Michael Galkin summarizes several more exciting recent papers applying diffusion models to molecular graphs. Check out the post for more detail, but some highlights:
Diffusion models for molecular conformer generation
Diffusion for molecular docking
Diffusion for structure-based drug design and generation of novel ligands
And finally, dive into a comprehensive Twitter thread for more:

Scientists are Using AI To Dream Up Revolutionary New Proteins (Ewen Callaway, Nature)
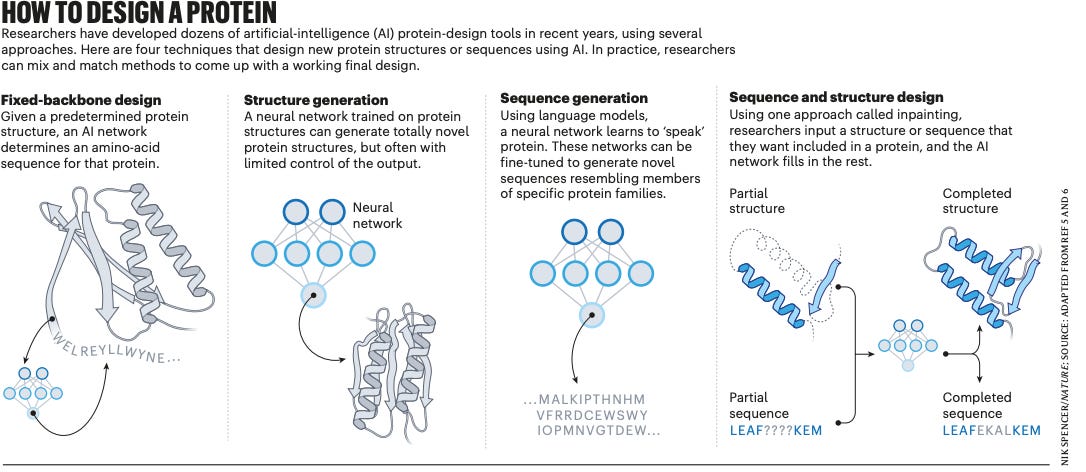
Using AI for novel protein design is likely not a new concept to any of you. While the methods vary, the basic concept involves creating an amino-acid scaffold to a desired protein structure through trained neural networks and then filling in the sequence and scaffold with the rest of the protein. A major concern has long been how to actually predict the right flow from sequence to structure, but advances in AI have deemed this issue solvable. What’s more interesting is the realm of possibilities that open to us once we nail the science of generating novel proteins, and of course, the associated scientific unknowns. An interesting thought experiment lies in food. Can we design entirely new sources of protein for human consumption? What allergy and immunogenicity ramifications might this give rise to? De novo protein design allows us to build with biological building blocks without a comprehensive understanding of the underlying biology.
Academic papers
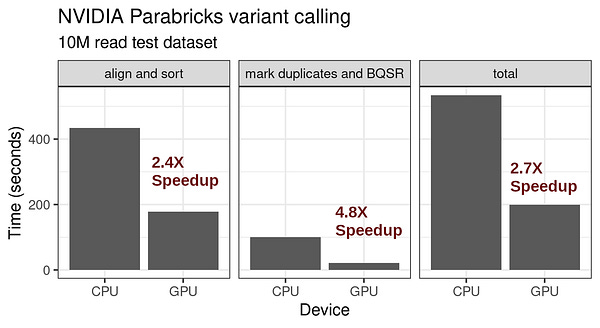
NMR-guided directed evolution [Bhattacharya et al., Nature, 2022]
Millions of years of evolution have engineered enzymes that synthesize molecules more quickly and efficiently than human chemists are capable, a phenomenon Derek Lowe refers to as “enzyme envy”. In an effort to replicate evolution to optimize enzymes with a desired or optimized function, scientists use a protein engineering method called directed evolution that involves an iterative process of 1) creating a library of protein variants using mutagenesis, 2) selection of variants with a desired function, and 3) amplification of selected variants for the next cycle.
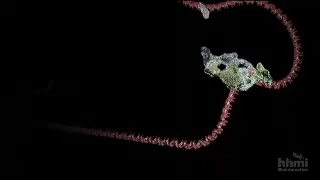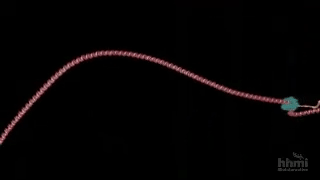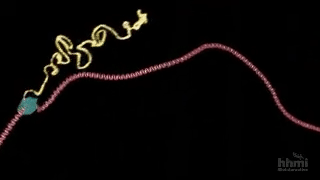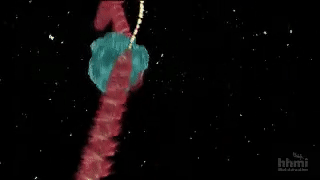
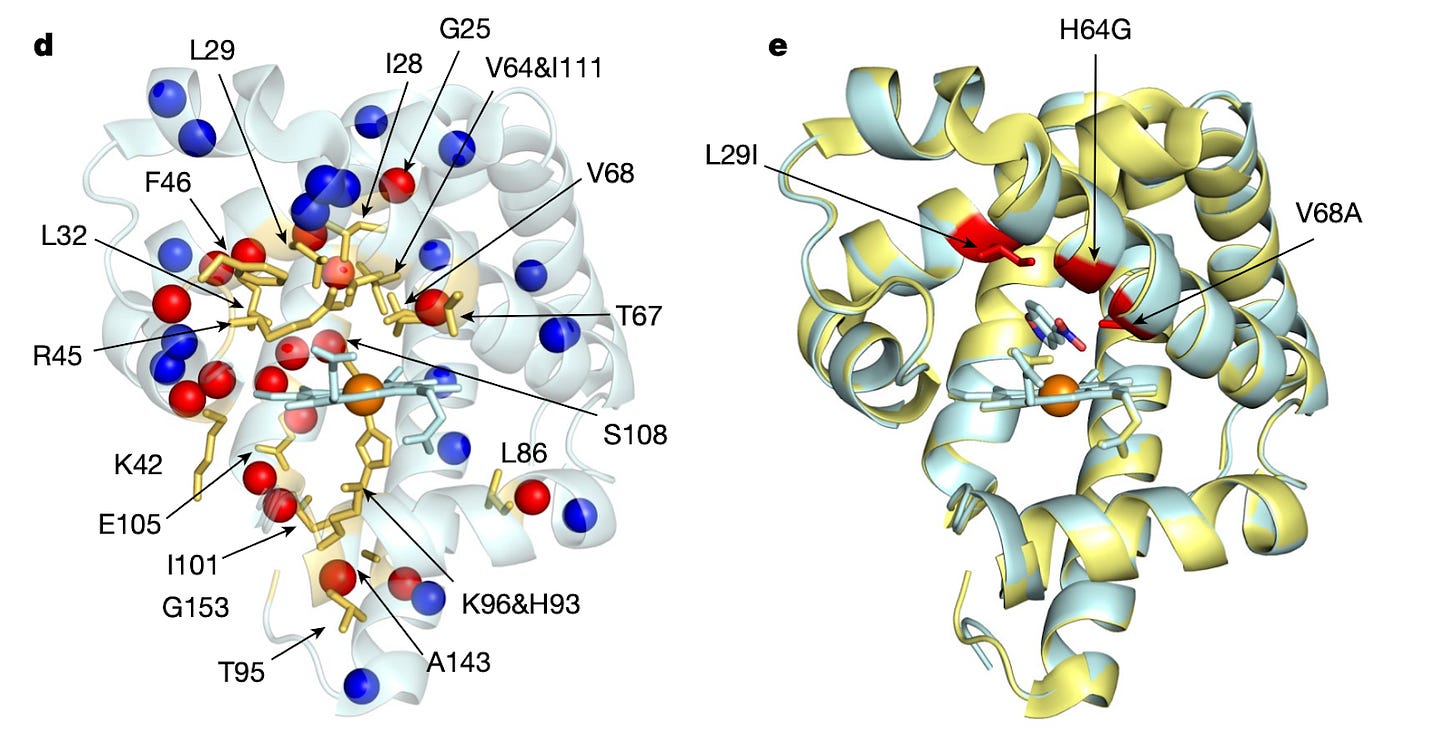
Despite its power, directed evolution is a brute-force method, and therefore becomes computationally intractable as the size of the protein or search space increases. Sampling the full sequence space of a 100 amino acid protein would require generating 20^100 variants, far too many to characterize experimentally. So the name of the game is narrowing down the protein sequence search space to be explored by predicting specific mutations that will result in the desired function of interest. Nuclear magnetic resonance imaging (NMR) is a useful technique for characterizing single amino acid residue information without requiring full structural characterization of the protein. Bhattacharya and colleagues adapt NMR to quickly predict productive mutations for the design of new proteins. Starting with the muscle protein myoglobin, the method was capable of identifying hot spots for mutation (yellow sticks in the left side of the figure below) and converted myoglobin into a highly efficient Kemp eliminase enzyme using only 3 mutations (red in right of figure). Given the minimalist and inexpensive approach (no structural and bioinformatic information is required), this has the potential to become a scalable, general-purpose method for engineering proteins via directed evolution.
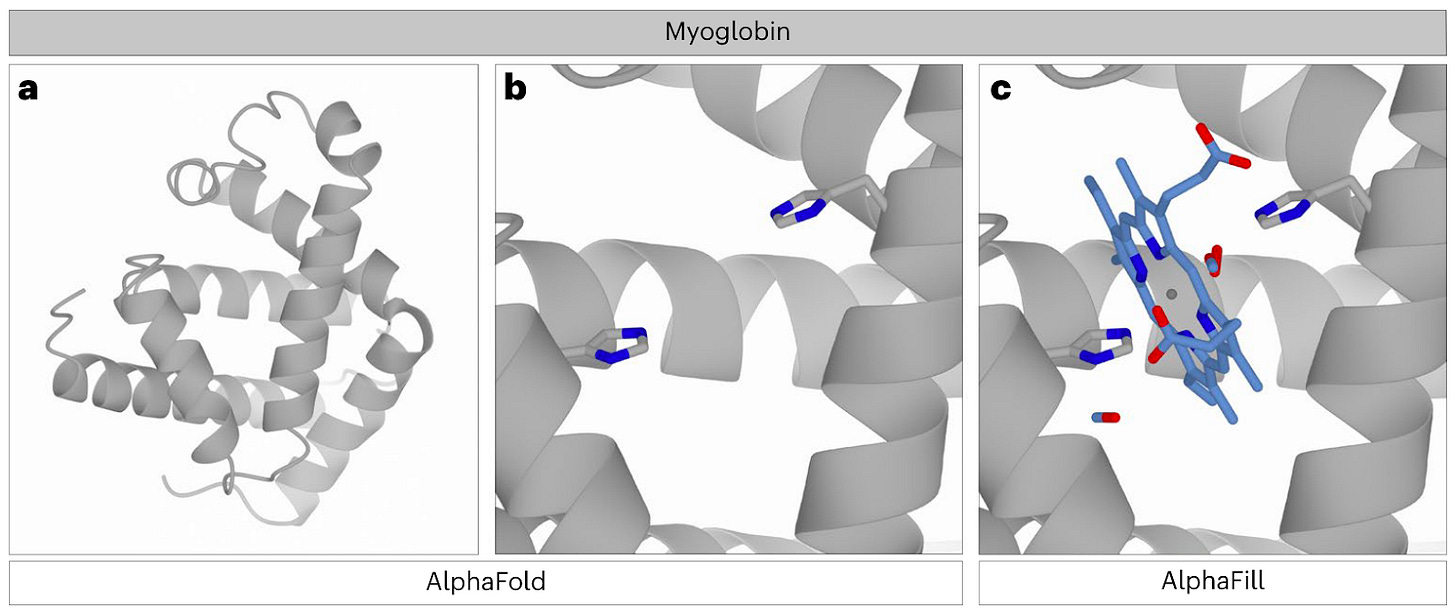
AlphaFill: enriching AlphaFold models with ligands and cofactors [Hekkelman et al., Nature Methods, 2022]
Why it matters: understanding the contacts of proteins to cofactors, ligands and ions, helps understand both the function and structural integrity of proteins, which can also be useful when designing downstream experiments.AlphaFold does not include small molecules, such as ligands and cofactors, in its models. For instance, zinc ions in zinc finger motifs or heme in hemoglobin are essential for structural integrity or function. AlphaFill completes the AlphaFold structural models by ‘transplanting’ ligands from experimentally-derived structures from the PDB-REDO database. However, the AlphaFill algorithm does not limit the ‘transplanting’ to the same protein, but extends it to structural homologs of the protein.
APOE4 impairs myelination via cholesterol dysregulation in oligodendrocytes [Blanchard et al., Nature, 2022]
Why it matters: Welcome to the post-amyloid hypothesis world! Though it is well-understood that carrying one copy of the APOE4 gene variant increases one's risk for Alzheimer's disease threefold (two copies about tenfold), the underlying pathophysiology of Alzheimer's in APOE4 patients has remained largely unknown. Researchers at MIT leveraged scRNAseq techniques to characterize how oligodendrocytes mismanage cholesterol in patients with one or more copies of APOE4.Leveraging postmortem human brains and lab-based human brain cell cultures, researchers at MIT generated a scRNAseq data asset featuring more than 160,000 individual cells of 11 different types from the prefrontal cortex in APOE3 and APOE4 patients. The initial analysis demonstrated that APOE4-carrying oligodendrocytes (neuroglia cells that provide insulation to the axons of the central nervous system) exhibited greater expression of cholesterol synthesis genes and disruptions to cholesterol transport.
Further exploration of the tissue in APOE4 brains showed excess cholesterol accumulation in the cell bodies of oligodendrocytes. These excess internal fats showed signs of stressing endoplasmic reticula involved in cholesterol transport, resulting in external lipid transport dysfunction. APOE4 brains also showed less myelination around axons.
Inspired by cholesterol’s potential role in APOE4 brains, the team explored whether drugs that target cholesterol, such as statins and cyclodextrin, could address these findings. Though statins did not appear to impact APOE4 brain cholesterol, applying cyclodextrin to APOE4 oligodendrocytes (cultured in a dish) reduced cholesterol accumulation within the cells and improved myelination in co-cultures with neurons. This research materially informs new Alzheimer’s hypotheses, though we appreciate that understanding the pathophysiology in APOE4 patients will require a greater appreciation for the interplay among microglia, astrocytes, and neural vasculature.
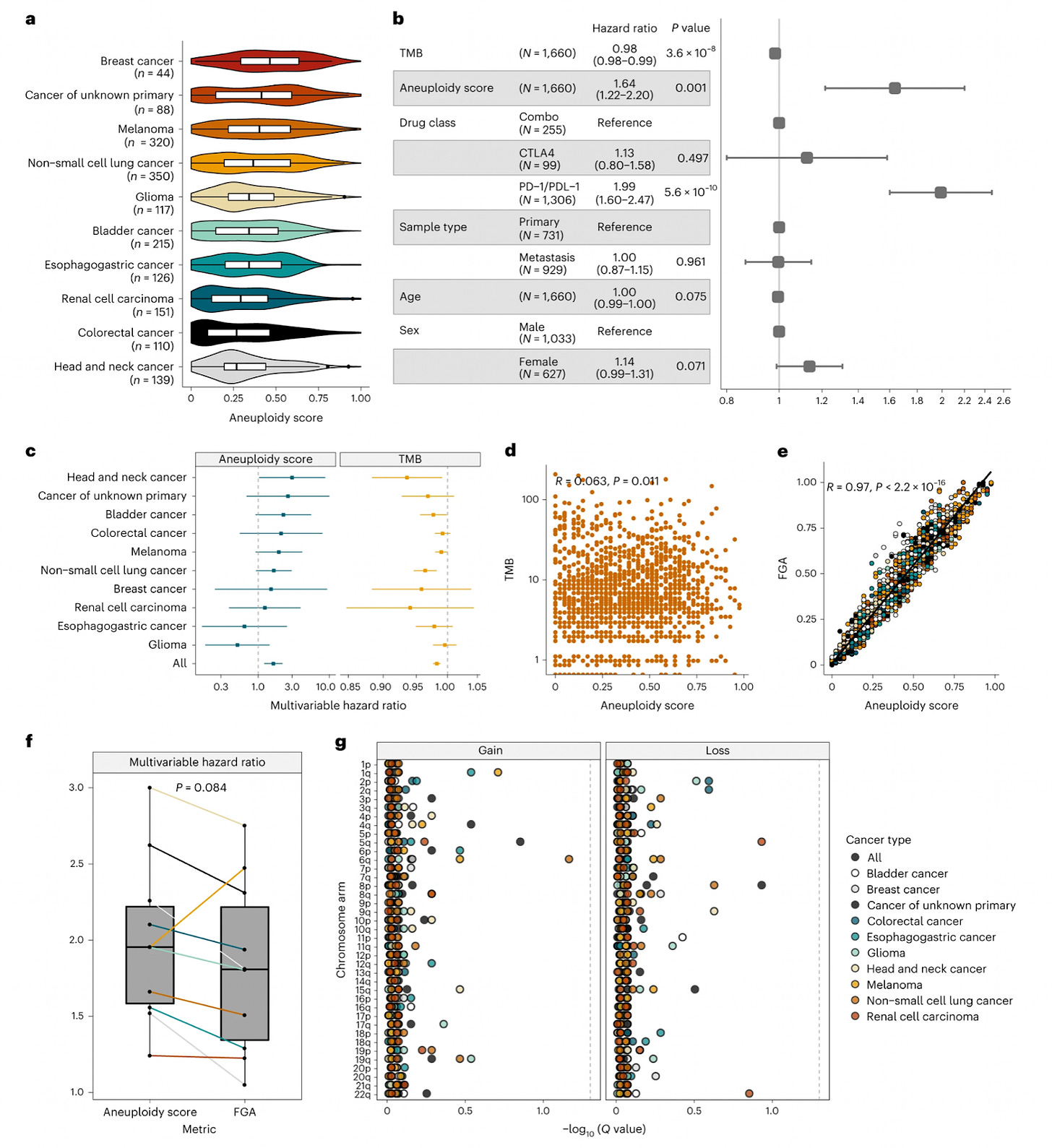
Tumor aneuploidy predicts survival following immunotherapy across multiple cancers [Liam et al., Nature 2022]
Why it matters: Although immunotherapy has changed the standard of care for many patients with late-stage cancers, it has remained challenging to identify those who will respond well to treatment. Over the years, several different biomarkers have been explored, with modest success therein. In this paper, Spurr et al., demonstrate that tumor aneuploidy (an unbalanced number of chromosomes/arms) is an independent prognostic value amongst patients with lower tumor mutational burden - specifically, amongst those with a low TMB, a higher aneuploidy score is associated with poor prognosis following immunotherapy.The authors reanalyzed a cohort of 1660 patients treated with immune checkpoint inhibitors, and found that aneuploidy was ubiquitous across tumors, while aneuploidy scores varied significantly by cancer type. Indeed, a higher aneuploidy score was associated with a worse prognosis, and was independently associated with overall survival across cancer types. Interestingly enough, specific chromosomal changes themselves (i.e. loss of heterozygosity of 9p21) were not associated with survival when controlling for aneuploidy score.
What we listened to
The Brain Inspired Podcast is one of the best podcasts covering the intersection of neuroscience and AI. In this episode, guests Evelina Fedorenko (a language neuroscientist at MIT) and Emily Bender (a computational linguist at UW) and host Paul Middlebrooks discuss the similarities and differences between language models and human language, studying language in animals, the interface between language and thought, and other topics.
Sam Harris speaks with Siddhartha Mukherjee about the human desire to understand and manipulate heredity, the genius of Gregor Mendel, the ethics of altering our genes, the future of genetic medicine, patent issues in genetic research, and other topics.
Notable Deals
Segmed Raises $5.2M Funding Round and Launches Real World Imaging Platform. Segmed is a cloud-based imaging platform for aggregating and providing access to healthcare data. Segmed forms partnerships with healthcare systems and aggregates medical data with the goal of delivering high-quality, real-world medical imaging datasets to clinicians, researchers, and developers building clinical AI systems. The round was led by Nina Capital, with participation from iGan Partners, M3 Inc, Mighty Capital, Expeditions Fund, and Alchemist Accelerator, bringing total funds raised to more than $10 million.
AI-enabled patient monitoring platform care.ai scores $27M in funding. Care AI has developed AI-enabled sensors for ambient sensing and the collection of real-time data for clinical and operational insight in healthcare environments. Applications include infection prevention, patient monitoring and fall prevention, and workforce optimization. The $27M round was led by Crescent Core Advisors.
Strand Therapeutics Announces Series A1 Bringing Total Round to US$97 Million. Programmable mRNA company Strand has added an additional $45M to its Series A financing round, bringing the total amount raised in the Series A to $97 million. New investor FPV led the round, with participation from Eli Lilly and Company, Potentum Partners, and existing investors Playground Global, and a further unannounced syndicate.
Cajal Neuroscience Launches with $96 Million to Transform Target and Drug Discovery in Neurodegeneration. Cajal is uniquely focused on the mechanistic, spatial and temporal complexity of neurodegeneration, integrating deep expertise in neuroscience, neuroanatomy and computational biology with state-of-the-art technologies for high-throughput functional validation. The financing was led by The Column Group and Lux Capital, with additional participation from Two Sigma Ventures, Evotec, Bristol Myers Squibb, Alexandria Venture Investments, Dolby Family Ventures and other investors.
What we liked on Twitter






Field Trip
The Casino and the Genie, The Generalist

Did we miss anything? Would you like to contribute to Decoding TechBio by writing a guest post? Drop us a note here or chat with us on Twitter: @ameekapadia @pablolubroth @patricksmalone @morgancheatham @ketanyerneni















Yet another great review! Love reading these.