BioByte 013
Scientific style, Galactica crashes and burns, a framework for trustworthy AI, frame editing, multi-omic single-cell fate mapping, TYK2, Horizon, and more!
Welcome to Decoding TechBio, a writing collective focused on the latest scientific advancements, news, and people building at the intersection of tech x bio. If you’d like to connect or collaborate, please shoot us a note here. Happy decoding!
Welcome to the December middlegame. Hope you have your holiday recipes all planned out, and don’t forget – calories don’t count if you’re not counting. We’d also avoid giving family and friends a dish you’ve cooked for the very first time, unless you want to star in the 2023 rendition of Kitchen Assassination. Safe travels those of you beginning the annual winter exodus to family and friends.
What we read
Blogs
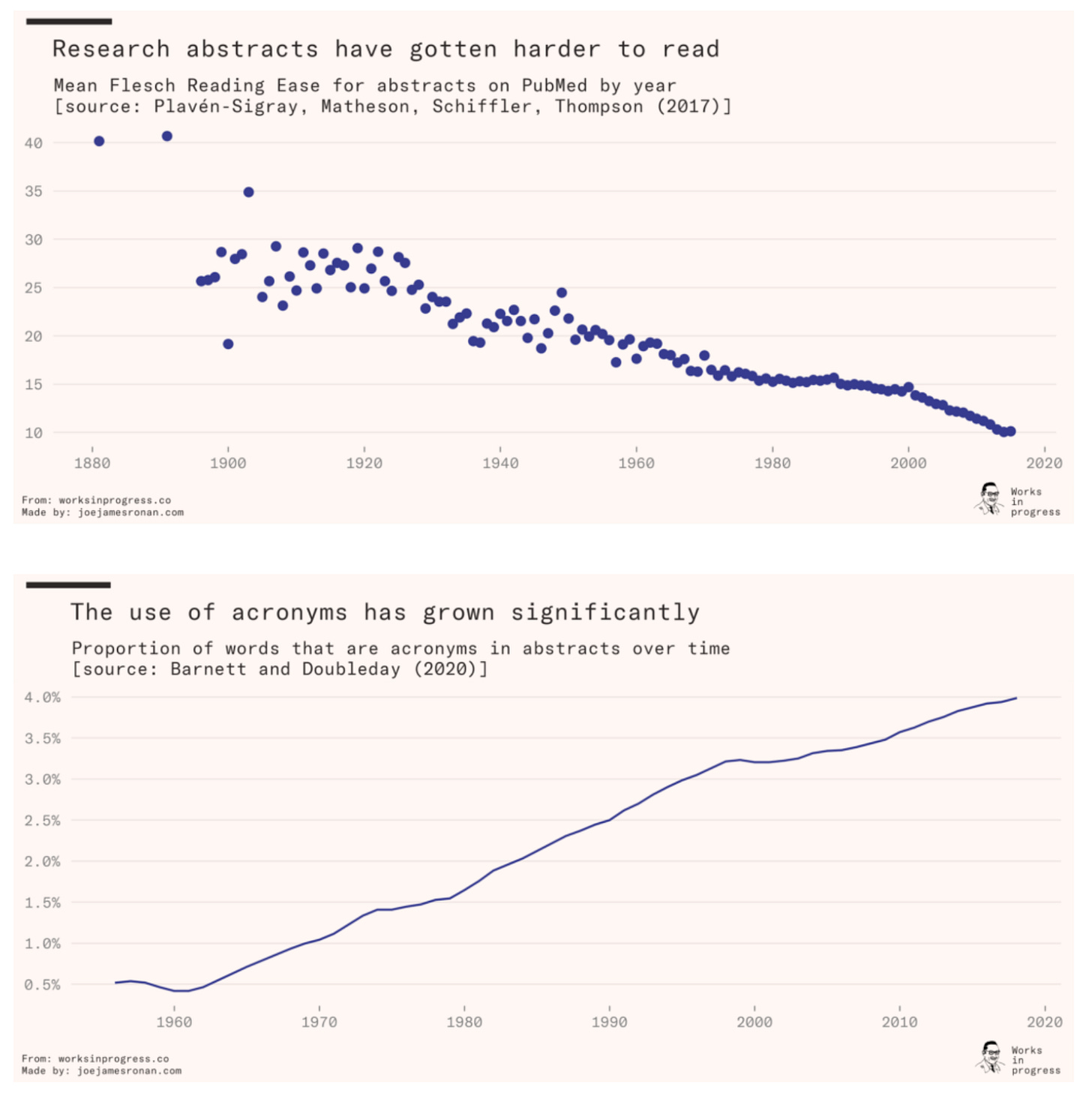
The elements of scientific style [Étienne Fortier-Dubois / Works in Progress, 2022]
Why it matters: It is difficult, if not impossible, to keep up with and digest the latest scientific papers. There are a bunch of reasons, but a predominant one is that the quality of scientific writing is declining. Étienne Fortier-Dubois puts the problem in context, and proposes some solutions.
This piece is perhaps a tad out of place in this newsletter, but being proponents of scientific communication, we thought it appropriate to include a recent Works in Progress essay on scientific writing. Scientific papers are more numerous, dense, technical, and jargon-laden than ever before. At the same time, many journals have constrained word counts so more information is packed in fewer lines. Each line is punctuated with several citations, each a potential rabbit hole that interrupts the reader’s flow.
The piece summarizes the evidence that science is becoming less readable (figures below). The reasons are various, but in summary - scientists are not incentivized to write clearly. What can the scientific community do to improve the quality of scientific writing? A few solutions are discussed:
Interactive papers published with companion Jupyter Notebooks that allow the reader to rerun and reproduce analyses and figures
AI-based tools for assisting scientific writing like Writefull
Alternative journal formats such as Seed of Science or Distill (which is now on hiatus)
Writing papers like a story with a narrative
Hiring professional writers that assist large research teams communicate their work
Ultimately, science is practiced by human scientists that operate according to incentives. Publications are the currency of academic science, and it is often the case that quantity of papers trumps quality. Until scientists are rewarded with grants and tenure for communicating their work effectively, the quality of scientific writing may not improve.
Why Meta’s latest large language model survived only three days online [Will Douglas Heaven, MIT Technology Review, 2022]
Language models are not really knowledgeable beyond their ability to capture patterns of strings of words and spit them out in a probabilistic manner. It gives a false sense of intelligence.
Galactica is a large language model for science, trained on 48 million examples of scientific articles, websites, textbooks, lecture notes, and encyclopedias released by Meta on November 15th 2022. After three days of its release, and constant criticism, the big tech company took it down.
What happened? There were two major problems: firstly, meta touted Galactica as a reliable tool for researchers, and secondly Galactica could not distinguish truth from falsehood. When these two are combined, biased and false explanations can seem “right and authoritative” according to Michael Black, director at the Max Planck Institute for Intelligent Systems in Germany, who works on deep learning.
We think Julian Toglius, Associate Professor at NYU, sums it up in his tweet:
My considered opinion of Galactica: it's fun, impressive, and interesting in many ways. Great achievement. It's just unfortunate that it's being touted as a practical research tool, and even more unfortunate that it suggests you use it to write complete articles.
Academic papers
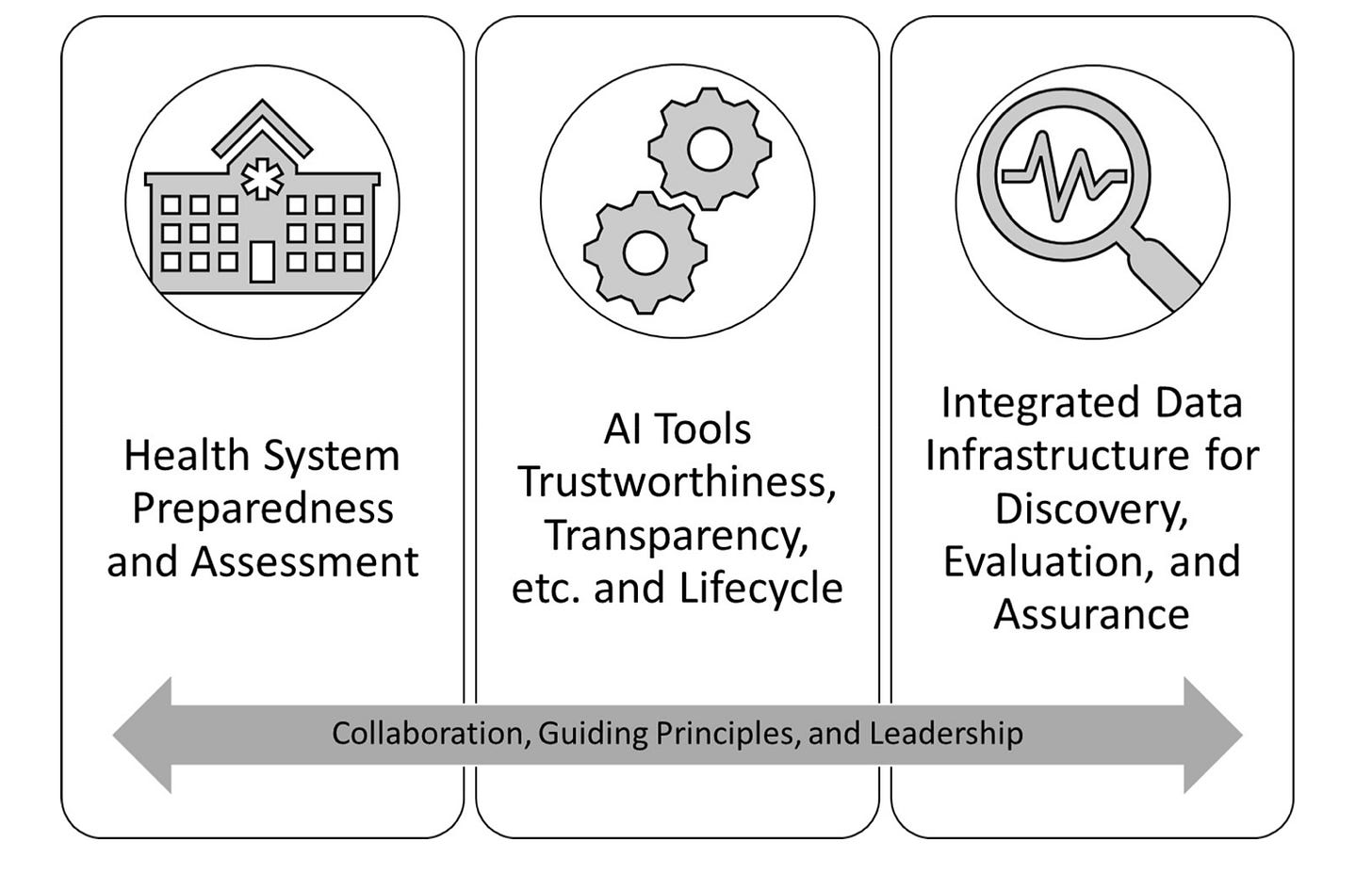
Blueprint for trustworthy AI implementation guidance and assurance for healthcare [Coalition for Health AI, 2022]
Why it matters: Despite strong technical progress in healthcare AI, real world impact for patients has lagged. There is a growing appreciation for the potential for AI to increase harmful outcomes and bias in healthcare. Better frameworks and tools are needed for ensuring the robustness and fairness of clinical AI. To this end, last week a group of government agencies, private companies, and medical centers formed a group called the Coalition for Health AI to advocate for such changes.The Coalition for Health AI published a white paper outlining a series of suggestions to improve the reliability and transparency of clinical AI systems, and included participation from Google, Microsoft, Optum, Mayo Clinic, Stanford, UCSF, the FDA, and the NIH. Several key elements for trustworthy AI in healthcare were defined:
Bias, equity, and fairness: AI should not exacerbate differences between groups, such as race or ethnicity. There should be multiple checkpoints for evaluation and monitoring of AI systems to account for changes in algorithm performance as a function of changes in the population or user behavior.
Testability: testability refers to how an algorithm’s performance is validated. Testing should be done through the algorithm’s life cycle, from the training phase through post-deployment. Testing is a continuous process, not a discrete step.
Usability: includes factors such as the quality of the user experience, both for providers and patients. Usability is heavily dependent on the context (e.g., clinical workflow considerations), and patient and provider perspectives should be incorporated into system design as early as possible.
Safety: an algorithm should first do no harm. Important to characterize safety risks of AI, such as automation bias.
Transparency: precise, standardized reporting of model details and performance from dataset curation to model design to final performance.
Reliability: An implementation guide should be included with every AI system to optimize workflow integration and prevent failure.
Monitoring: ongoing surveillance of AI to monitor for data drift, changes in user behavior, or other variables that can cause a change in algorithm performance. Central reporting should be completed so that all clinical sites can learn from the experiences of others.
To address these issues, the group called for the creation of an independent accreditation lab, where health systems and AI tool developers will submit their algorithms for evaluation. The organization would certify an algorithm’s use on a task so that proper uses and side effects can be disclosed. A registry for clinical AI, similar to clinicaltrials.gov, was proposed. None of these proposed changes are set in stone, but instead are meant to jump start a national discussion amongst stakeholders to drive greater focus on the real-world implementation of healthcare AI.
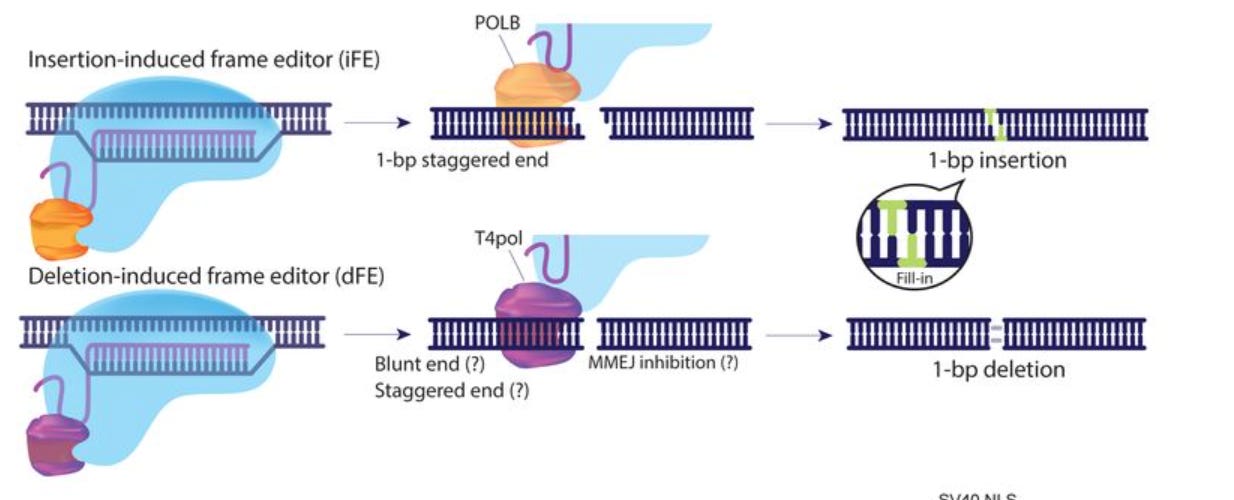
Frame Editors for Precise, Template-Free Frameshifting [Nakade et al., bioRxiv, 2022]
Following the coverage of PASTE last week, today we’re covering Frame Editors (FEs), a new Cas-based fusion editing system.
Teams from MIT and Hiroshima University published a preprint outlining a system to improve both editing efficiency and accuracy of CRISPR-Cas systems by fusing Cas9 with DNA polymerases. FEs can introduce precise frameshifts into target loci, to both insert (iFE) and delete (dFE) 1-base pair at a time.
When compared to Cas9, both types of FEs reduced the number of random mutations at target loci. Another advantage to FEs is that they don’t require donor template sequences. These donor template strands, which are commonly used to introduce frameshift mutations, tend to impair editing efficiency.
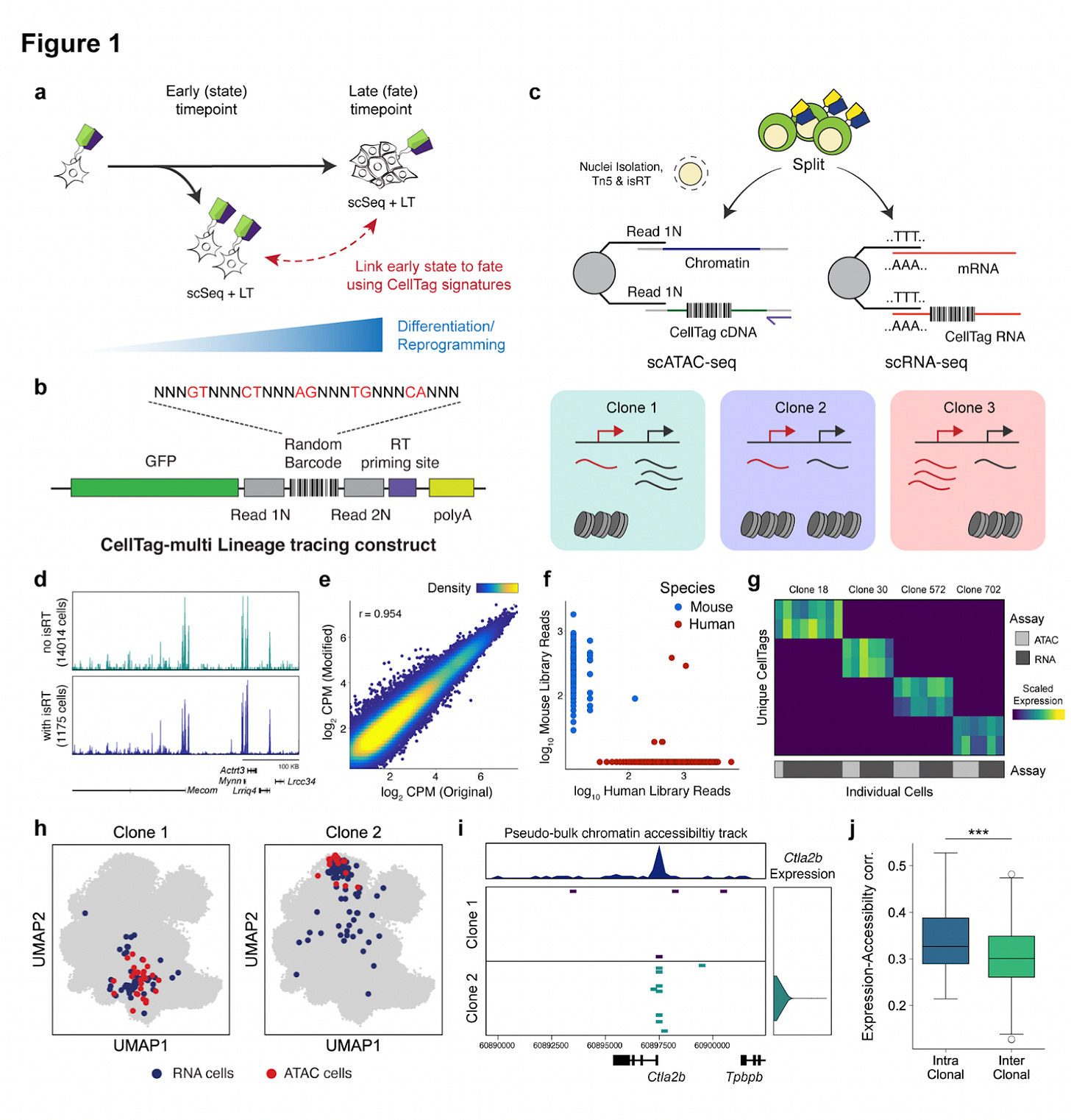
Why it matters: the single base pairs introduced or deleted by FE can correct frameshifts either by causing the mutation to revert to the normal genotype or by making the mutations small and in-frame. This technique expands the range of genetic diseases caused by frameshift mutations that could be cured. This is also an important step towards genome editing without templates.Multiomic single-cell lineage tracing to dissect fate-specific gene regulatory programs [Jindal et al. bioRxiv, 2022]
Why it matters: For years, scientists have been trying what “omics” modality can be used to best understand and predict cellular developmental fate, especially during hematopoiesis. Recent work has shown that transcriptomics has low predictive accuracy for such fate mapping. In this paper, Jindal et al., show that multi-omic single-cell lineage tracing reveals that transcriptional and epigenomic information is significantly better at predicting cell fates. Here, the authors develop CellTag-multi, which uses sequential lentiviral delivery of heritable random barcodes, CellTags, allowing multi-level lineage tree construction. These CellTags are polyadenylated transcripts that can be captured via both scRNA and scATAC-seq, enabling both transcriptional and epigenomic state tracking. By applying CellTag-multi in lineage reprogramming of fibroblasts to endoderm progenitors (iEPs), they reveal hidden chromatin remodeling dynamics, and identify certain transcription factors (TFs) that influence cell fate. Indeed, only the addition of multi-omic profiling uncovered the importance of certain TFs in gene regulatory network dynamics.
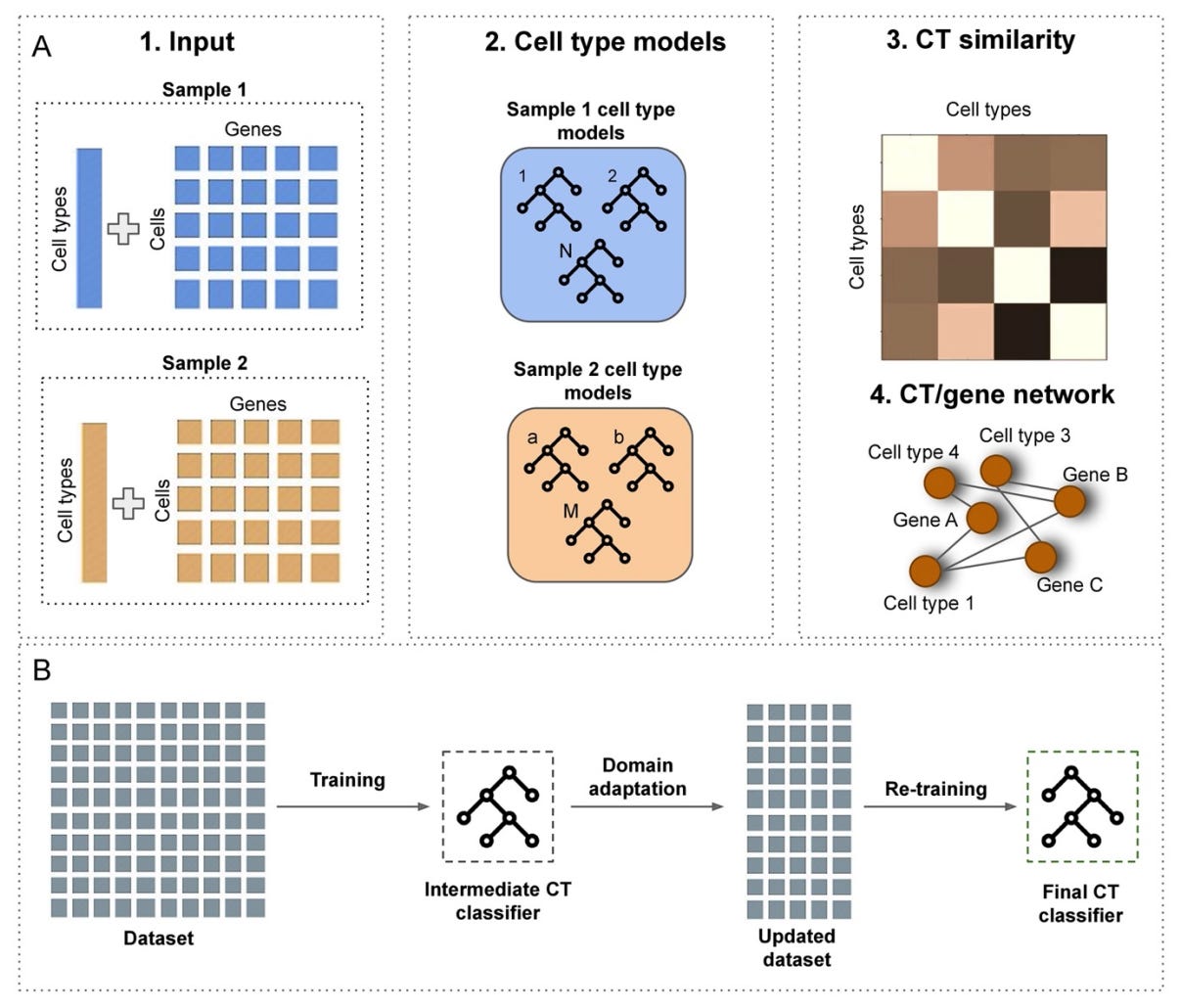
scEvoNet: a gradient boosting-based method for prediction of cell state evolution [Kotov et al., bioRxiv, 2022]
Why it matters: Understanding the developmental history of cell states offers insight into the molecular characteristics of various cell types, and from a therapeutic perspective, can help researchers understand tumor-to-metastasis transitions. Though scRNA-seq methods enable understanding of gene expression characteristics of a given cell state, there is currently a dearth of computational tools that can articulate the evolution of cell states, such as how cell states change molecular profiles (e.g., novel gene activation).This paper by Kotov et al. proposes scEvoNet, a gradient-boosting-based method for the prediction of cell state evolution in cross-species or cancer-related scRNA-seq datasets. scEvoNet works by building a confusion matrix of cell states and a parallel network connecting genes and cell states, which allows users to obtain genetic data shared by a common cell state even across distantly-related data sets (think cross-species). Within the network, these genes can be interpreted as markers of evolutionary divergence or tumor evolution in tumor-to-metastasis transitions. One limitation of the method is that scEvoNet does not match gene sequences, instead only leveraging user-provided labels, which might reduce the number of genes to be found across cell types in cross-species analyses. Take scEvoNet for a spin here!
What we listened to
Hear from Michal Preminger, head of JnJ innovation and Jacob Plieth, Evaluate Vantage journalist, as they discuss the latest in biotech.
Semil Shah opines on whether “partner” or “brand” matters when it comes to VC, and how Haystack and his own investment career came about.
Notable Deals
Takeda Acquires Nimbus’ Tyk2 Autoimmune Program. Nimbus Therapeutics, a clinical-stage company that designs and develops breakthrough medicines through its powerful computational drug discovery engine, announced that it has signed a definitive agreement under which Takeda will acquire Nimbus Lakshmi, a wholly-owned subsidiary of Nimbus Therapeutics, and its tyrosine kinase 2 (TYK2) inhibitor NDI-034858. NDI-034858 is an oral, selective allosteric TYK2 inhibitor being evaluated for the treatment of multiple autoimmune diseases following successful recent Phase 2b results in psoriasis. For a trip down memory lane, check out Atlas Venture’s original blog post on Nimbus here.
Former BridgeBio exec launches new anti-inflammatory biotech with $169M. Paragon Therapeutics, an antibody engineering company, has spun out its anti-inflammatory antibodies into Apogee Therapeutics, which launched out of stealth Wednesday morning with a Series B of $149 million. At the helm is former BridgeBio chief business officer Michael Henderson, whose former company imploded late last year after its lead amyloidosis drug failed a Phase III trial. All in all, the new anti-inflammatory biotech has $169 million to work with. It’s keeping quiet on what exactly it’s working on, though Henderson did offer up a timeline on when Apogee’s programs would be in the clinic.
Amgen to buy Horizon Therapeutics for nearly $28B in cash. Amgen will be bringing on two blockbuster drugs, helping buffer themselves against the competition from the biosimilar space. At $116.50 a share, Amgen is paying a 48% premium over Horizon’s price. Horizon boasts $3.6B in revenue (driven mainly by Tepezza, for thyroid eye disease), while projecting that Krystexxa (for gout) will hit $1.5B a year in revenue.
In case you missed it
How AI That Powers Chatbots and Search Queries Could Discover New Drugs
What we liked on Twitter







Events
WuXi Global Forum 2023 - Advancing Breakthroughs for Patients | January 10, 2023 | SF
WuXi will be hosting a half-day event at JPM covering:
The State of Innovation 2023
Breaking Barriers: Druggability Redefined
Opening New Horizons: Spotlight on RNA Therapeutics
Leading Next Generation of Cell and Gene Therapies
and more!
Field Trip
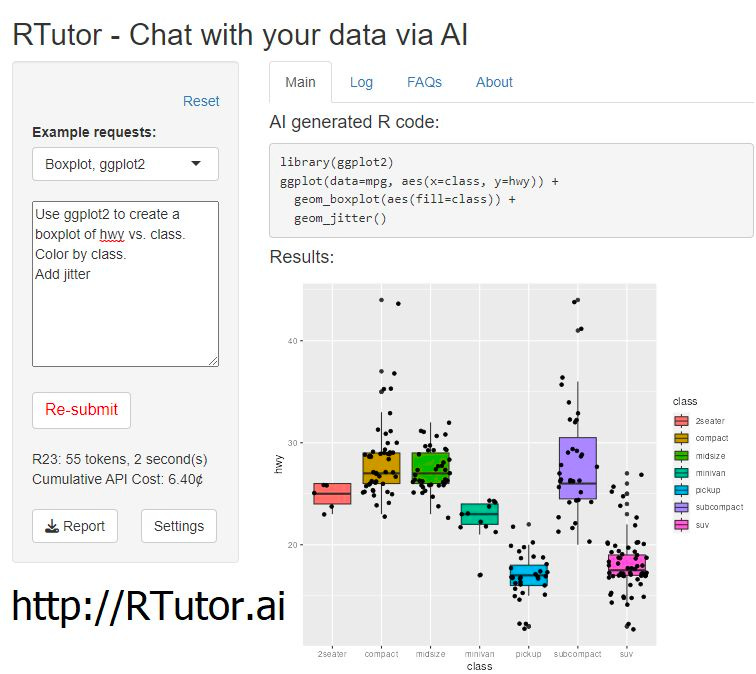
Did you know you can build a virtual machine inside of ChatGPT?
RTutor launched yesterday - "an application that can generate and test R code by chatting with it, based on the powerful Davinci (ChatGPT's sibling) from @OpenAI.”
Did we miss anything? Would you like to contribute to Decoding TechBio by writing a guest post? Drop us a note here or chat with us on Twitter: @ameekapadia @pablolubroth @patricksmalone @morgancheatham @ketanyerneni