BioByte 027: RL for multi-step chemistry, transcriptomic profiling for neurological diseases, the upending of cortical organizational theory, new funding models for biomedical innovation
Welcome to Decoding Bio, a writing collective focused on the latest scientific advancements, news, and people building at the intersection of tech x bio. If you’d like to connect or collaborate, please shoot us a note here or chat with us on Twitter: @ameekapadia @ketanyerneni @morgancheatham @pablolubroth @patricksmalone. Happy decoding!
The year is 2047. A biosimulation of Frank Ocean returns to Coachella and performs the entirety of Channel Orange for a crowd of late 40’s / early 50’s millennials. Nature heals.
We’re still iterating on a bunch of experiments here at Decoding Bio and appreciate the community’s continued support. Starting this week, we’re going to begin gathering and sharing insight from the Decoding Bio community.
We’ll share the results next week. Happy (belated) DNA Day!
What we read
Blogs
AI system can generate novel proteins that meet structural design targets [Adam Zewe, MIT News, 2023]
Researchers from MIT, the MIT-IBM Watson AI Lab, and Tufts University published a paper showcasing the results of an attention-based diffusion model when trained to design de novo proteins.
Attention-based models can learn long-range relationships, required to develop proteins due to the long-distance effects of a single amino acid change on the overall structure of the protein. Diffusion models enable the generation of high-quality new data based on design objectives.
The design objectives were based on secondary protein structures. These produce different mechanical properties: proteins with alpha helix structures yield stretchy materials while those with beta sheets yield more rigid materials. The model inputs include a desired percentage of secondary structure components and then generate amino acid sequences for these. This is connected to AF2 or OmegaFold which predicts the 3D structure of the protein.
The proteins have yet to be experimentally validated, but the team validated that the model was outputting reasonable proteins by inputting physically impossible design targets, but were impressed to see that instead of producing improbable proteins, the models generated the closest synthesizable solution.
Initial use cases for the model, the authors mention, include developing protein-inspired food coatings, keeping produce fresh longer while being safe for consumption, novel materials with certain mechanical properties, and replacement materials for petroleum-derived objects.
The State of Computational Protein Design [Alissa Hummer, Oxford Protein Informatics Group, 2023]
Oxford PhD student Alissa Hummer wrote up a short summary of the Keystone Symposium on Computational Design and Modeling of Biomolecules, and the state of the protein design field. Unsurprisingly, deep learning methods such as RFDiffusion from the Baker lab were one of the most important topics. Despite advances in ML-based methods like RFDiffusion and ProteinMPNN, it is important to note that physics/energy-based methods like Rosetta are still commonly used, and in fact, many state-of-the-art systems combine both approaches.
The most interesting takeaway is the trend toward conformational flexibility being increasingly incorporated in computational protein design. Proteins are dynamic, shape-shifting structures, which can be difficult to model computationally. There are several groups developing methodologies to address this, such as the labs of Tanja Kortemme and Anum Glasgow who developed methods for modeling conformational flexibility from allosteric interactions in the lac repressor system. Other unsolved problems in the space that were discussed included the role of water molecules in protein function, and important gaps in knowledge of the biophysics governing conformational flexibility and water-protein interactions.
Why biotechs should buy more of their software [Kaleidoscope, 2023]
Kaleidoscope is a new software for bio companies creating project and asset management, automation, and workflow tracking for biotech companies. The team has summarized learnings in working with biotech companies across various functions to understand gaps in tooling and infrastructure. Currently, many biotechs build out specific internal tools and software products based on their needs. In an industry where the need is very specific to workflow, data type, and use case, not to mention additional regulatory specifications, it’s been difficult to find custom software solutions to adopt. The problem with building internal tooling is that it ends up looking a lot like running a software company within a biotech company—two totally different structures that are only now starting to show some common threads in terms of organizational structure and focus. Kaleidoscope lays out three major takeaways from their conversations to build a case for why internal tooling is putting a bandaid on a deeper problem of needing better R&D software products:
Time-and-money-intensive: Building internal software is expensive, both in terms of time and money to create a usable tool, and in terms of the complexity in maintaining that tool over time.
Expensive indirect costs: As an R&D company, your valuable resources should be used to advance your core IP; building internal tooling is a big distraction from doing the science that matters.
Adapting existing software: While your workflows may feel very unique, the abstracted problems are much more ubiquitous. Modern external software can get you 90% of the way and be adapted for the last mile, for far less time and money, and for a far better product experience.
On the other hand, you can argue that the needs of biotech companies are diverse enough that an off-the-shelf solution isn’t viable for all your workflows and won’t scale well as the company’s R&D efforts evolve. It’s hard to deny that better software is a key issue as biotech evolves to incorporate more and more complex data but a suite of products is likely not a standalone holy grail solution. This industry in particular will require more customization per product than typically seen with software. It’ll be interesting to see how enterprise solutions meet R&D teams in the middle.
Academic papers
AlphaFlow: autonomous discovery and optimization of multi-step chemistry using a self-driven fluidic lab guided by reinforcement learning [Volk et al., Nature Communications, 2023]
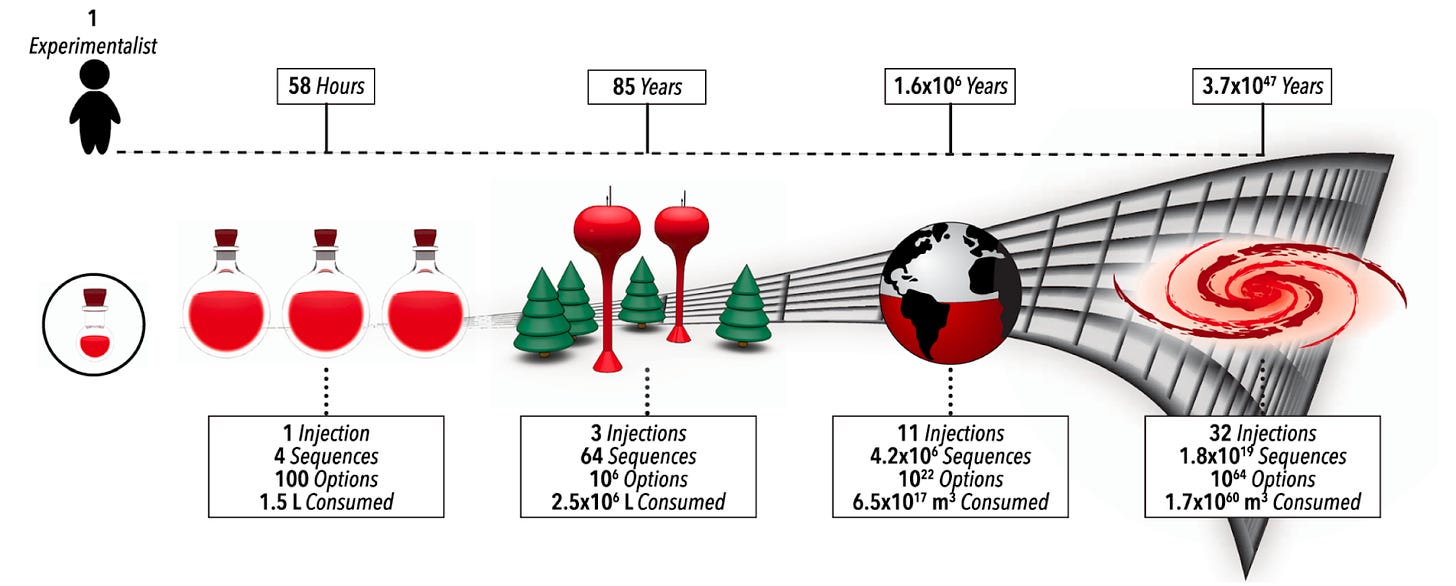
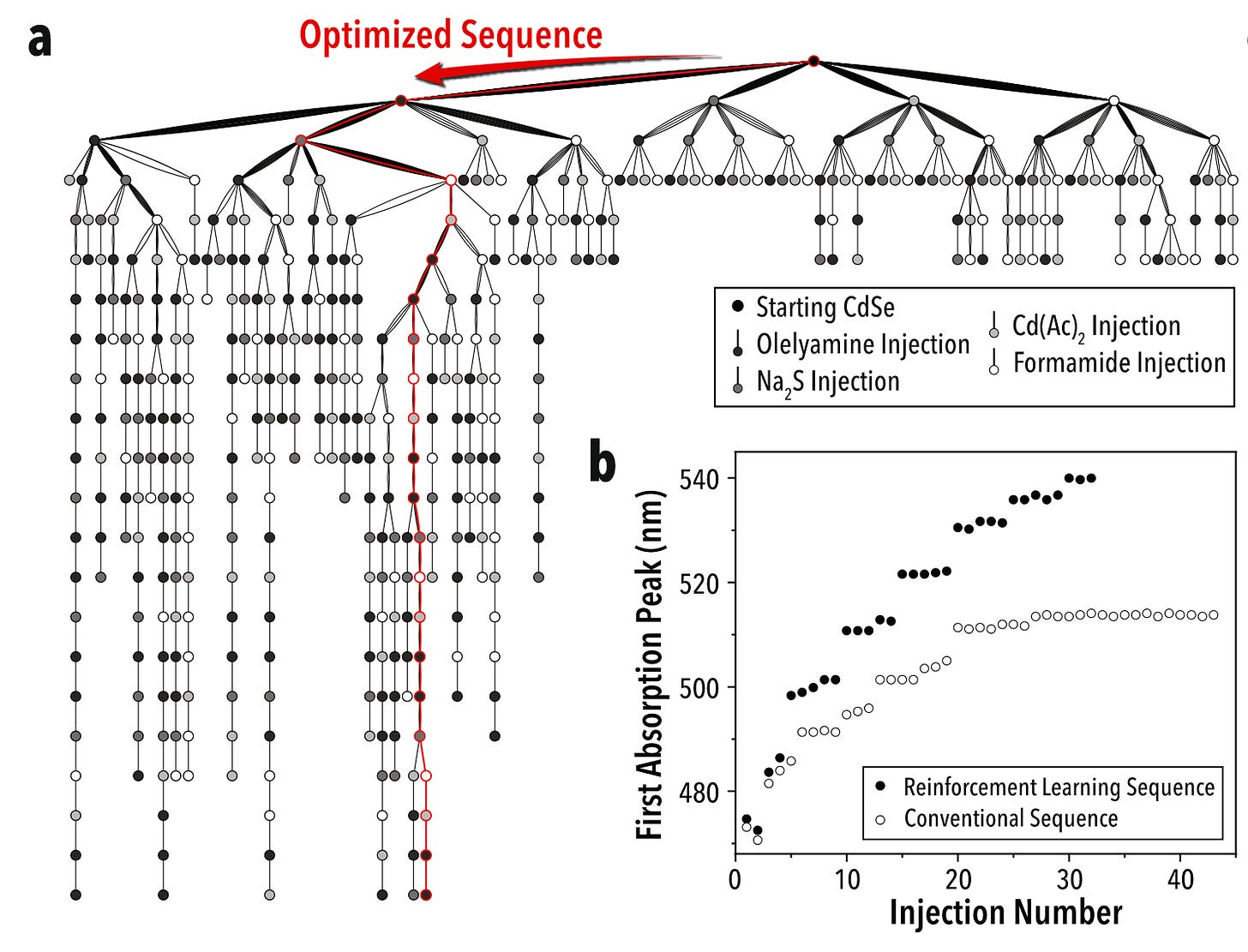
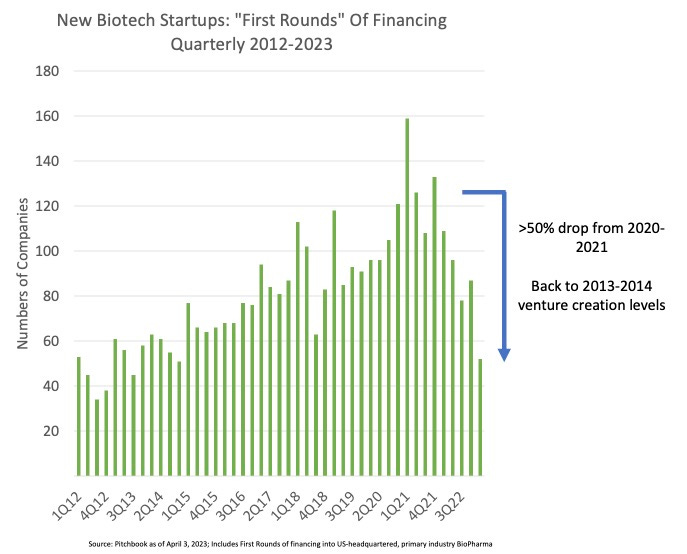
Why it matters: For self-driving labs (SDLs) to become a widespread option for multi-step chemistry, they must overcome two challenges: dimensionality and data scarcity. With each step in a reaction, the number of parameters grows exponentially (as seen above), and therefore also does the cost and time required to find optimal synthetic routes. This team managed to overcome both with this platform.Following last week’s dive into LLM-powered lab automation, today we talk about how these researchers from North Carolina State University and University at Buffalo automated the discovery and optimization of complex multi-step chemistry using a self-driven, reinforcement learning-led fluidic lab.
In an analogous way to AlphaGo, AlphaFlow uses reinforcement learning to optimize reagent quantity, volume, and other parameters to generate the best result. In the paper, the authors use their self-driving lab for the precision synthesis of heterostructure quantum dots using colloidal atomic layer deposition. This is a perfect application given the large number of steps and parameters in the multi-step reaction (its complexity exceeding 40 dimensions).
The platform ran miniaturized and automated experiments, and through trial and error identified the best synthetic route as measured, in this case, by a higher absorption peak wavelength. AlphaFlow indeed found a better synthetic route that is commonly utilized and optimizes reaction conditions to increase the quality of the nanomaterial.
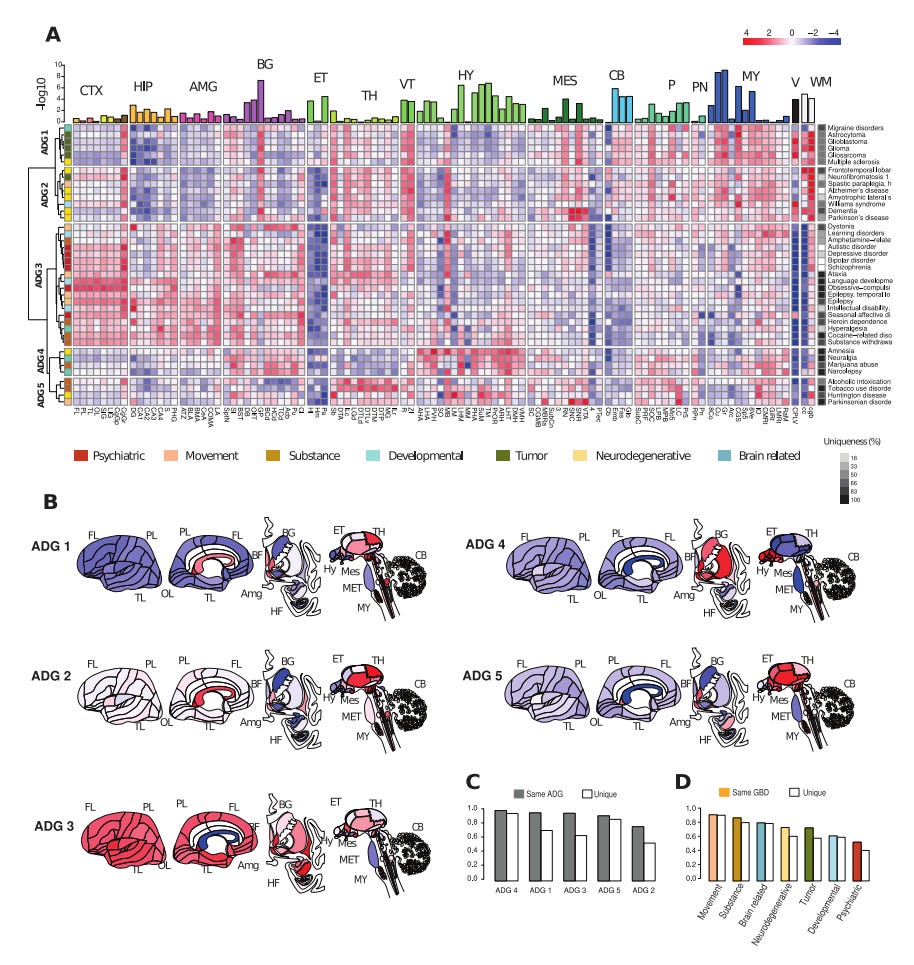
A comparison of anatomic and cellular transcriptome structures across 40 human brain diseases [Zeighami et al., PLOS Digital Biology, 2023]
Why it matters: Disease phenotyping is one of the most exciting applications of -omics data in biomedicine today. These efforts are particularly compelling in indication spaces such as neurology where lagging understanding of underlying pathophysiology has held back progress in developing curative therapies. A recent analysis of 40 common human brain diseases used transcriptomic data to identify and localize major transcriptional patterns in adult brains, offering a molecular-based strategy for classifying and comparing neurological diseases and potentially, novel disease relationships.This study demonstrated spatial and temporal manifestations of neuropathology by investigating transcriptomic relationships leveraging the Allen Human Brain Atlas and six human post-mortem brains. Mean gene expression profiles for genes associated with 40 major brain diseases (including Alzheimer's, Parkinson's, ALS, epilepsy, addiction, autism, bipolar disorders, and frontotemporal dementia) were characterized across >100 anatomic structures spanning the cortex, hippocampus, amygdala, basal ganglia, and more. Single-cell analysis across homologous cell types in both mouse and human samples validated disease risk genes in common cell types while preserving species-specific expression and phenotype. Ultimately, the study results produced a molecular classification of neurologic disease based on five groups: 1) tumor-related, 2) neurodegenerative, 3) psychiatric, and 4) substance abuse, and 5a/b) two mixed groups of diseases affecting basal ganglia and hypothalamus.
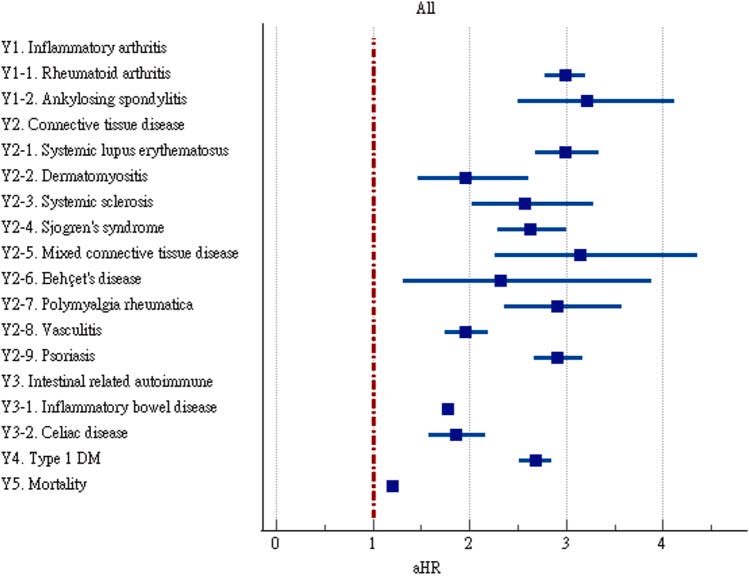
Heightened risk of autoimmune diseases after COVID-19 [Sharma & Bayry, Nature Reviews Rheumatology, 2023]
Why it matters: there’s an emerging link between viral infections and onset of autoimmune disease that’s difficult to study given the long timelines between the two. New cohort studies suggest a link between COVID-19 and risk factor for autoimmunity.We’ve previously covered emerging links between viral infections and the onset of certain autoimmune diseases (for example, Epstein-Barr virus and multiple sclerosis). Longitudinal cohort studies are difficult to run and are polluted with confounding factors, hence the lack of understanding of mechanisms and links between viruses and autoimmunity. Two recent large cohort studies of COVID-19 patients show there may be some increase in risk factor for autoimmune disease following COVID-19 infection. It’s hard to tell whether this risk factor is because of suppressed/heightened immune response to the virus itself, T-cells response that went berserk, residual inflammation, or more likely a combination of these factors and more. These cohorts of patients were also unvaccinated as the way in which vaccination confounds results is unknown. Unsatisfyingly, the conclusion here is we need to do more work to uncover relationships between autoimmunity and viral infection, specifically on the mechanism of action and triggers.
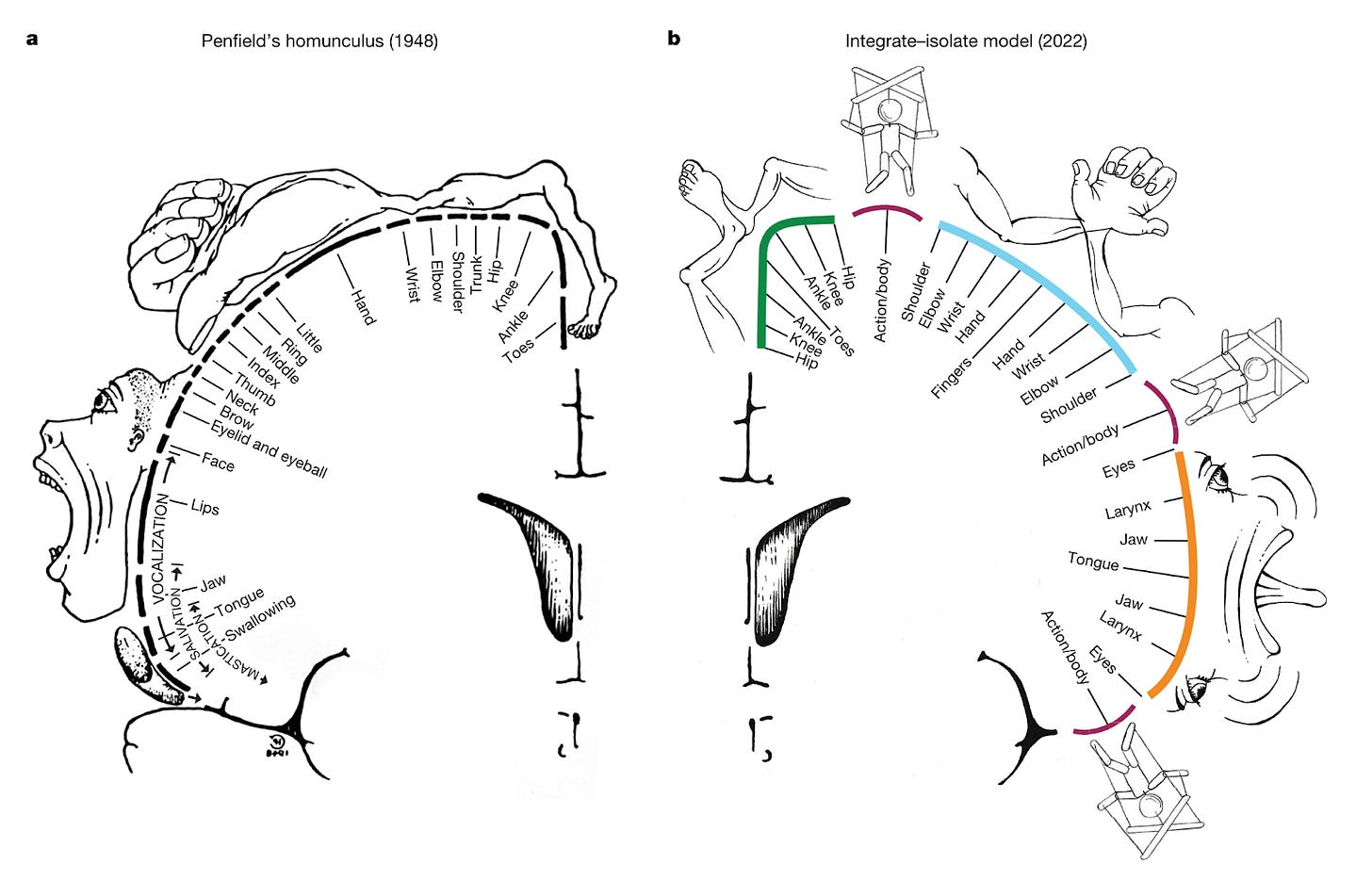
A somato-cognitive action network alternates with effector regions in motor cortex [Gordon et al., Nature, 2023]
Why it matters: A top neuroimaging lab upends the textbook theory of primary motor cortex cortical organization!If you’ve taken a neuroscience course, you’ve probably seen the images depicting the somatotopic arrangement of the primary motor cortex (aka M1, see figure left below). The textbook view for decades has been that specific subregions of M1 control different parts of the body; the dorsal portion of M1 on top of your head controls your legs, the more lateral portion closer to your ears control your arms and hands. In a landmark paper in Nature this week, Evan Gordon and colleagues from one of the top neuroimaging labs in the world provide compelling evidence that revises this model.
The authors used a method called precision fMRI to functionally segment the primary motor cortex into different subregions. Precision fMRI uses a combination of functional connectivity (i.e., correlating fMRI time series for different brain regions as a proxy for connectivity), task-based fMRI, and structural MRI to map individual-level brain organization. They found that the classic homunculus in M1 is interrupted by sub-regions with distinct patterns of connectivity and function that are conserved across adults, kids, babies, and even macaques. These subregions are highly interconnected within M1, but also connect to higher-level executive function networks involved in action planning, but movement itself. Confirming these findings, a task-based experiment showed that these regions were more involved in action planning than execution. This new network is termed the somato-cognitive action network (SCAN).
There are many implications for these findings, but one of the most interesting is for the design of brain-computer interfaces. Motor prostheses are the OG application of BCIs, and there are now several startups entering or soon to enter clinical trials for paralysis (Synchron, Neuralink, among others). These devices were designed with the classic model of M1 in mind. Less consideration has been given to a subnetwork within M1 like SCAN that is responsible for action planning for goal-oriented movement. For BCIs to continually improve quality-of-life for patients with movement disorders, additional research will be needed to learn the neural code for this network.
What we listened to
Scientist Stories: Andrew Lo, New Funding Models for Biomedical Innovation
Notable Deals
European VCs raise fresh funds for life sciences investing: Forbion and Gilde Healthcare have raised $750 million and $600 million euros, respectively.
Enveda Biosciences announces oversubscribed Series B1 bringing total round to $119 million
In case you missed it
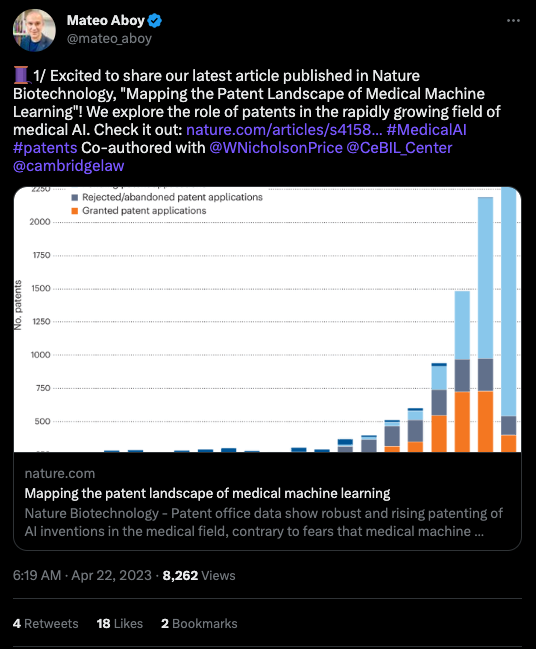
What we liked on Twitter
As a reminder, Substack is no longer supporting Twitter previews. Click the image to visit the tweet!
Events
Field Trip
It’s allllll about Kaytranada this week!
Did we miss anything? Would you like to contribute to Decoding Bio by writing a guest post? Drop us a note here or chat with us on Twitter: @ameekapadia @ketanyerneni @morgancheatham @pablolubroth @patricksmalone