BioByte 051: mapping the human body, foundation models, effective fundraising and rational molecular glue design
Welcome to Decoding Bio, a writing collective focused on the latest scientific advancements, news, and people building at the intersection of tech x bio. If you’d like to connect or collaborate, please shoot us a note here or chat with us on Twitter: @zahrakhwaja @ameekapadia @ketanyerneni @morgancheatham @pablolubroth @patricksmalone. Happy decoding!

Your weekly dose of TechBio. Just in time for a rainy weekend.
Feedback time: please shoot us a message on what you have been liking and what you’d like to see done differently… we’re always keen to hear your thoughts
What we read
Blogs
Mapping the Human Body [Ian Hay, Positive Selection, October 2023]
One of the primary reasons 90% of clinical trials fail is a disconnect between lab research, which focuses on studying cells and molecules in isolation (bottom-up), and clinical medicine, which focuses on treating patients' symptoms and diseases (top-down). In a new essay, Ian Hay proposes a new framework to help bridge this divide, in which AI is used to model individual cells and their interactions which lead to emergent system-level effects. In this framework, a model represents each cell as an artificial neural network that takes in data on the cell's gene expression and outputs how the cell will change its behavior by modifying its gene expression. The neural networks are trained on actual gene expression data from single-cell RNA sequencing experiments. These individual cell models are then connected together to model cell interactions in tissues and organs. The model can simulate the effect of drugs or other perturbations by altering the gene expression of cells and seeing how this propagates through the system. As more cellular data is added, the model grows in scope, with the ultimate goal being a complete map of cell interactions in the human body that can predict the system-wide response to therapies. Importantly, such a model would connect cell-level and system-level biology, establishing more accurate prediction of whether therapies would translate to human patients.
Biotech milestones for effective fundraising [Kaleidoscope, 2023]
Friends and members of Decoding Bio share some pointers on what milestones they expect from biotech companies at different stages of financing in order to unlock further funding. The authors hope that by bringing some more transparency to the process, it can provide some rubrics as to what investors look for. Some quotes from the blog:
Data moats: “how data is used to drive decisions throughout the entire organization, not just on the R&D side. With respect to the R&D side, data-driven KPIs and metrics will be very company-specific. Some examples include: number of experiments per week, cost per experiment, cost per x parameter, cost per screen, amount of data generated, cost to IND, predicted value per asset, NPV for existing pipeline, predicted productivity of the platform. We diligence these factors consistently regardless of whether the opportunity is a new incubation, a Seed, Series A or a later stage company.”
Platform validation: “We usually look for validation of the platform. Which entails having run it at least once. For instance, if the company is optimizing viral vectors, we'd like to see that i) the company has built its proprietary in silico methodology and HT wet lab assay to screen vectors, ii) have run it and iii) those runs have yielded something that wouldn't have been possible without the platform. In this case, it could be that they found a capsid that has better tropism for a specific cell type. The metric by which success is measured will depend on the platform's objective (e.g. % higher tropism for a particular cell type if its a delivery platform, % higher affinity for a small molecule or % better bioavailability)”
Lead and target identification: “The primary things to strive to show are] identification of lead indication and target, preclinical data generated demonstrating target involvement in disease, and ability to modulate target to improve disease phenotype. The goal of Seed financing is to get a development candidate for the lead program before the Series A.”
Team and opportunity: “In the earliest stages, there isn't a whole lot to dig into. The big question is "is this a world-class team working on a massive problem with the right motivations and tech foundation" There's technical diligence to do on whether the approach to solving the problem makes sense, doesn't break laws of physics, and can scale but a lot comes down to the founders, their ability to hire and execute, and the market. We'll get into the specifications and economics of the technology and the go-to-market plan but at pre-seed and early seed, those things are almost always going to change so those questions are more to get a sense for how founders think about the opportunity than something to base all of diligence on.”
Check out the blog for more insights at different stages of financing.
The Foundation Model Transparency Index [Bommasani et al., Stanford, October 2023]
This paper by Bommasani et al. introduces the Foundation Model Transparency Index, a new methodology for assessing the transparency of major foundation model developers like OpenAI, Anthropic, and Google. The index evaluates developers across 100 indicators that comprehensively measure transparency across three domains: upstream (data, compute), model (capabilities, risks), and downstream (distribution, impact).
The index finds that transparency in the foundation model ecosystem today is minimal, with an average score of just 37/100 across developers. Upstream transparency related to data, labor, and compute is especially poor. Even factors like model size and architecture are often not disclosed. Downstream impact is almost completely opaque.
By scoring developers, the index incentivizes companies to improve their transparency over time. It also clarifies where transparency is most lacking, allowing policymakers to craft effective regulations. As more foundation models emerge for biology and chemistry, we are keen to see how frameworks such as transparency indices and industry-standard benchmarks emerge. Overall, the Foundation Model Transparency Index moves the AI field toward greater openness and accountability. See more here.
Platemaps aren’t hard but everything around them is [Jesse Johnson, Scaling Biotech, 2023]
In this essay, Jesse highlights a critical issue almost all lab scientists have run into—how to communicate experimental data (what’s in the plates) to the rest of the company, notably data teams. Tracking metadata and then actually using the metadata is a complex problem few companies have been able to do well. The reality is, this really shouldn’t be a complex problem. Metadata in the context of biotech startups is usually stored in some sort of table but somehow extracting that information and conveying it to the right people seems nearly impossible. Jesse traces the root cause of the problem to no standardization in the long, hand-wavy process of experimental design/editing. Software is archaic, experiments change overtime, the processes are one step too early for LIMS and electronic lab notebooks, etc etc. Jesse outlines the requirements of a solution to this problem and we’re aware of startups innovating to solve this obvious but important issue.
Academic papers
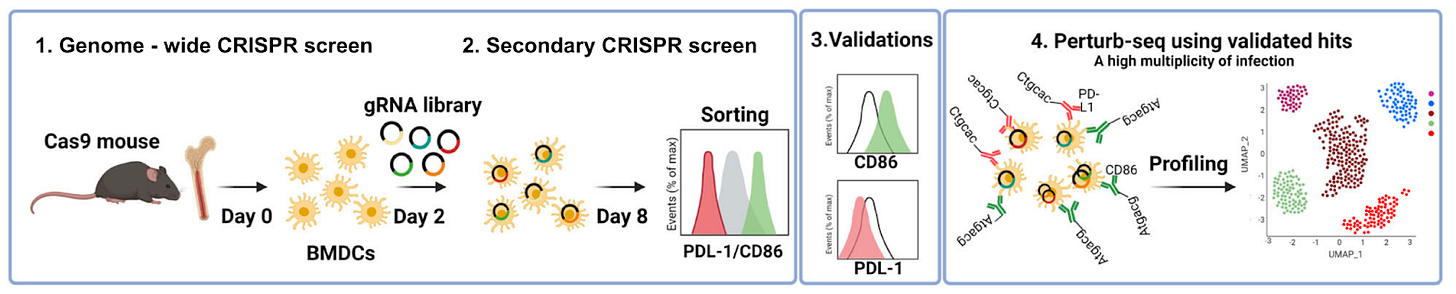
Systematic identification of gene combinations to target in innate immune cells to enhance T cell activation [Xia et al., Nature Communications, October 2023]
Why it matters: A new high-throughput functional genomics study identifies gene networks in dendritic cells that drive T cell activation and inhibition. By identifying gene-gene interactions that interact non-linearly to drive T cell behavior, combination therapies that tune immune cell function in disease may be developed. The activity of T cells in the human body is a delicate balance. For example, too much T cell activity can result in autoimmune disease. Too little, and a patient may not respond to cancer immunotherapy. Dendritic cells (DC) are antigen-presenting cells that play a critical role in T-cell priming. In this paper, pooled CRISPR screens were used to discover combinations of gene targets that rewire circuits in DCs to drive either T cell stimulation (assessed by up-regulation of CD86, a receptor expressed on DC's surface that drives T cell expansion) or inhibition (assessed by up-regulation of PD-L1, which inhibits T cell activation). Two genome-wide screens in DCs identified negative regulators of CD86, including Chd4, Stat5b, Egr2, Med12, and positive regulators of PD-L1, including Sptlc2, Nckap1l ,and Pi4kb. Finally, higher-order gene perturbation of gene combinations was performed by increasing the ratio of guide RNAs delivered via lentivirus (resulting in some cells with multiple edited genes), which identified synergistic gene-gene interactions.
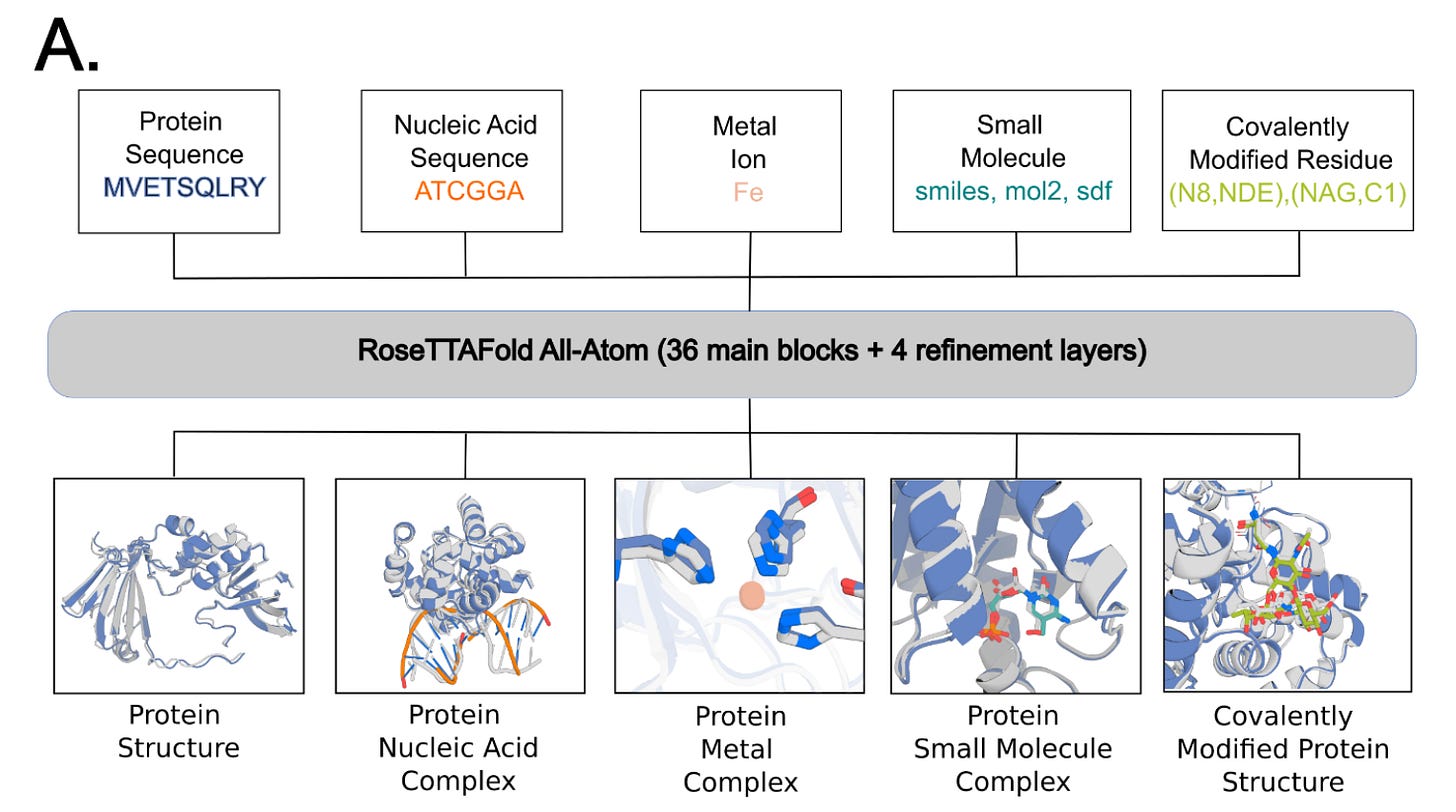
Generalized Biomolecular Modeling and Design with RoseTTAFold All-Atom [Krishna et al., bioRxiv, October 2023]
Why it matters: The deep neural networks AlphaFold2 and RoseTTAFold enable high-accuracy prediction of protein structures based on amino acid sequence. However, they are unable to model protein interactions with small molecules and other non-protein molecules (such as metals, nucleic acids). This is important to model for drug discovery but to also understand proteins in their natural environment which commonly interact with such types of molecules during cell signaling, metabolism, translation and transcription.This team from the University of Washington, which includes David Baker’s Lab, have developed RoseTTAFold All-Atom (RFAA), which is capable of modeling full biological assemblies. RFAA uses a combination of sequenced-based description of biopolymers (proteins and nucleic acids) with an atomic graph representation of small molecules and protein covalent modifications. This is distinct from the full atomic, LLM-based structure this team from U of Toronto/Vector Institute used to model small molecule and proteins together covered in BB046.
RFAA was trained using a protein-biomolecule dataset from the PDB, supplemented by small molecule crystal structures from the Cambridge Structural Dataset. Interestingly, the team used a technique they call “atomization” in which they feed portion of proteins as atoms rather than residues to augment the learning of general atomic interactions. In testing, RFAA predicted 42% of complexes successfully inCAMEO vs 38% by DiffDock (covered inBB024). Did someone say SOTA?
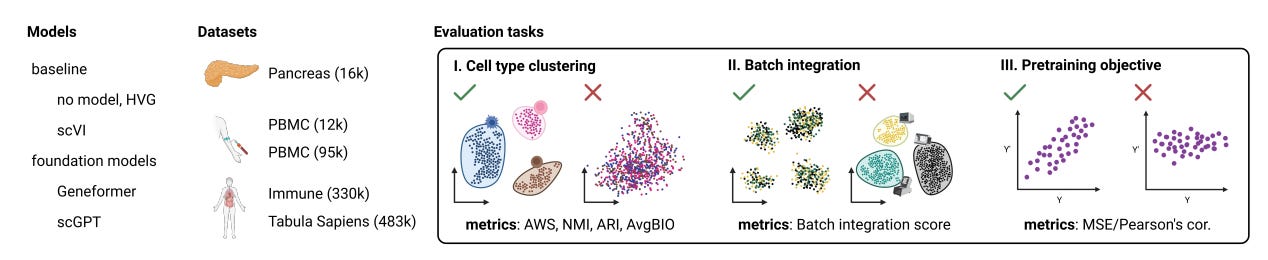
Assessing the limits of zero-shot foundation models in single-cell biology [Kedzierska et al., bioXriv, October 2023]
Why it matters: Foundation models hold promise to automate complex analytical workflows in single-cell biology. However, this study exposes limitations in adapting natural language models for biological data. It provides an important benchmark for future work on designing effective pre-training strategies and objectives tailored for single-cell transcriptomics. Rigorous zero-shot testing is key to developing truly generalizable models that biologists can deploy for exploratory analysis without additional training or tuning. More robust foundation models would accelerate discovery from single-cell datasets.The paper examines two recently proposed foundation models for single-cell transcriptomics - Geneformer and scGPT. Foundation models are pre-trained on large datasets to capture universal patterns, enabling adaptable reuse. The goal is for models like Geneformer and scGPT to serve as general tools for diverse single-cell analysis tasks.
The authors rigorously tested the models' capabilities when used in zero-shot mode, without any task-specific fine-tuning or training. They evaluated performance on clustering cell types, integrating batches, and reconstructing gene expression per the models' training objectives.
The results showed limited reliability in zero-shot settings. The models failed to outperform simpler techniques like selecting highly variable genes for clustering. For batch integration, they lagged behind existing methods like scVI. The models also struggled to accurately reconstruct gene expression, doing no better than predicting mean expression.
The findings indicate current single-cell foundation models lack robustness when applied out-of-the-box. The pretraining objectives may not teach meaningful biological representations. More focused research is required to realize the potential of foundation models for broadly generalizing across single-cell analysis settings.
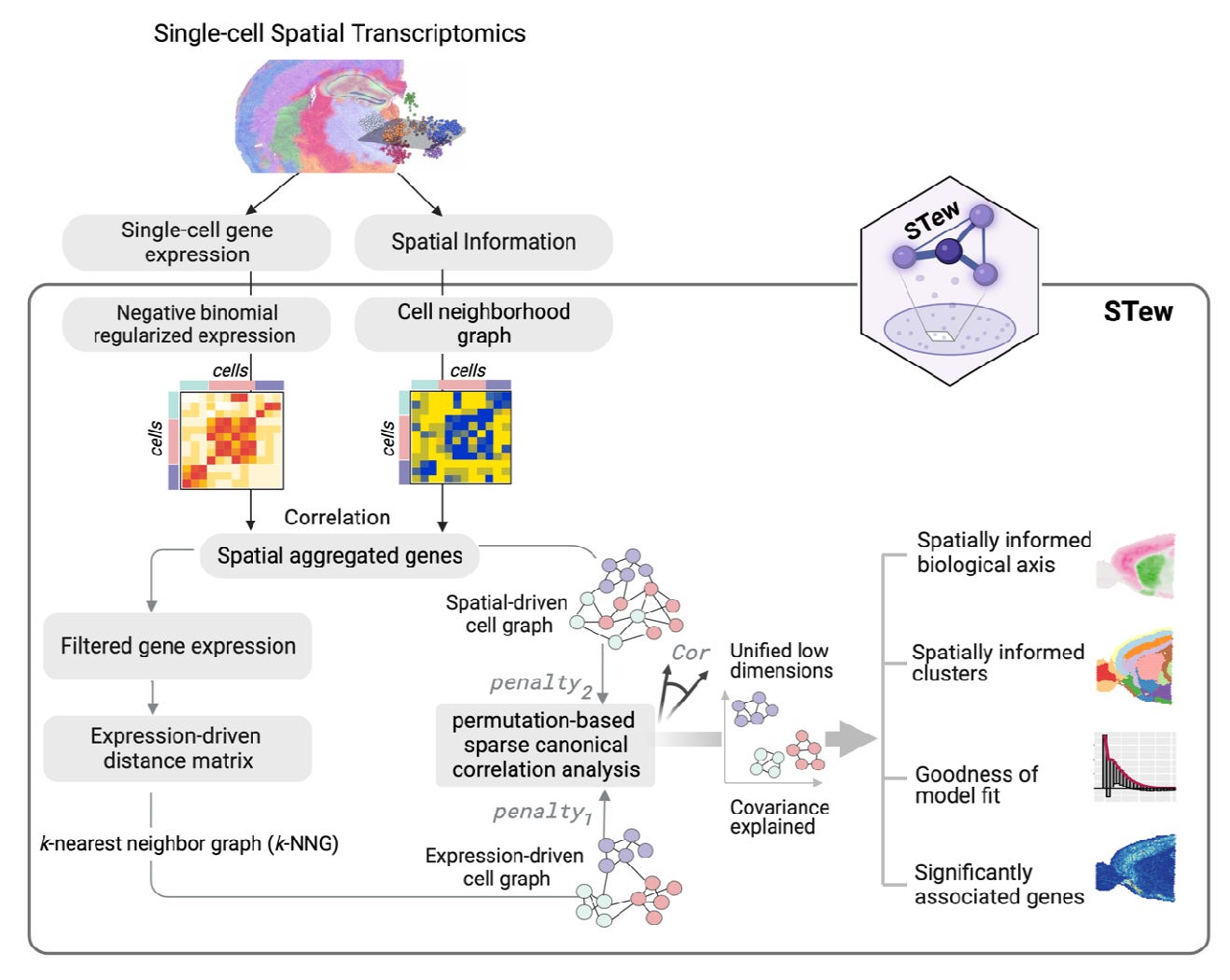
Uncover spatially informed shared variations for single-cell spatial transcriptomics with STew [Guo et al, BioXriv, October 2023]
Why it matters: Understanding how cells are spatially organized within tissues is key to elucidating tissue function in health and disease. Methods like STew that leverage spatial transcriptomics data will allow researchers to map diverse cell types and states to their locations. This can reveal how cellular neighborhoods and spatial context influence cell behavior and intercellular communication. These insights into tissue architecture could uncover new disease mechanisms and targets for therapy. More effective integration of spatial data with genetics has the potential to transform our understanding of tissue biology.The paper introduces a new method called STew for analyzing spatial transcriptomics data. Spatial transcriptomics technologies measure gene expression levels at defined spatial locations across a tissue sample, preserving information about where cells are located and their spatial relationships.
STew is designed to identify shared variations between the spatial locations of cells and their gene expression patterns. It uses graph-based methods to model spatial neighborhood relationships between cells. It then applies sparse canonical correlation analysis to find important shared features between the spatial graph and gene expression data. The authors tested STew on several spatial transcriptomics datasets from mouse and human tissues, including brain and skin. Compared to other methods, STew more accurately identified cell types and their spatial domains. It delineated smooth, continuous regions for each cell type within the tissue. STew also revealed biologically meaningful gradients that correspond to different tissue layers and structures. The authors added statistical modeling to identify genes significantly associated with the spatial patterns. This helps uncover subtle gene expression changes across tissue regions.
Notable Deals
Roche is Glue’d to Monte Rosa
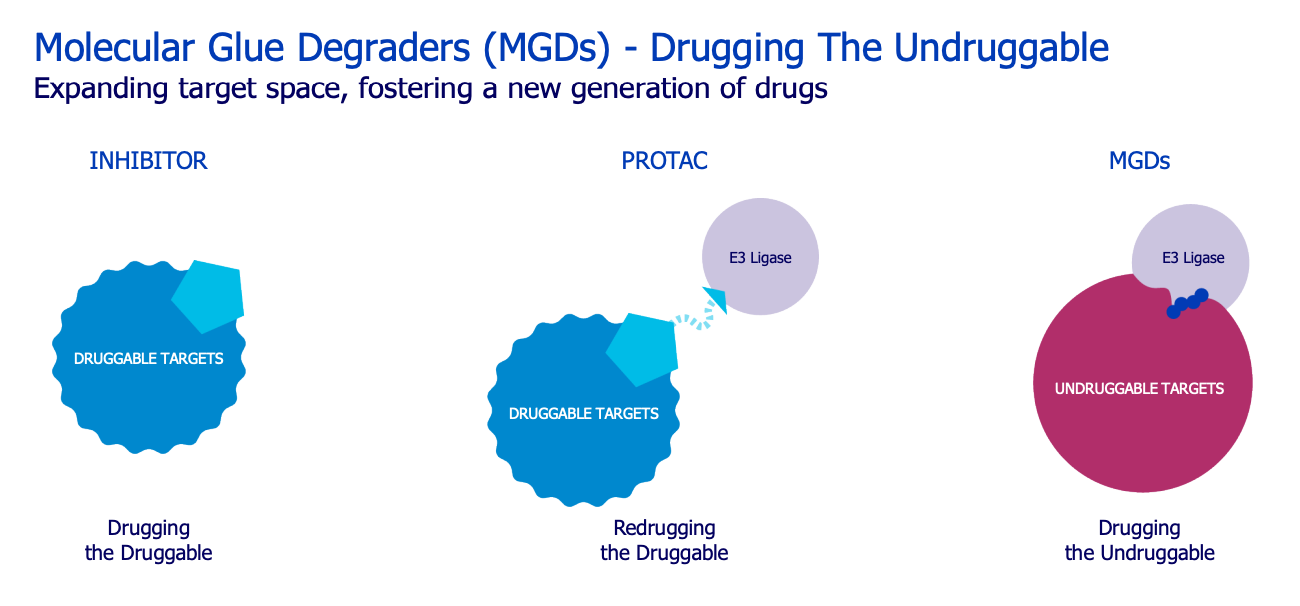
Monte Rosa ($GLUE): biotech focused on the development of small molecule targeted degraders called molecular glues (MGDs). These degrade the target protein by facilitating the protein’s interaction with an E3 ligase.
Deal: Strategic collaboration and licensing agreement to discover and develop novel a certain number of MGDs for cancer and neurological diseases. $GLUE will perform initial discovery and preclinical studies, after which Roche has the option to obtain worldwide exclusive license to the products.
Financials:
Upfront: $50M
Milestones: preclinical, clinical, commercial and sales milestones exceeding $2B, including $172M for achieving its pre-clinical milestones.
Additional targets: if Roche decides to include extra targets an it may pay an upfront of up to $28M and potential milestones of $1B
Royalties: $GLUE is eligible for tiered royalties from high single-digit to low teens on any commercialised products
Term: two years with option to expand at additional cost
Why did Roche partner with $GLUE?
Attractive modality: MGD’s are touted to unlock previously undruggable proteins since they do not require binding of the active site, but hook on to ‘degrons’. Many ‘holy grail’ targets have been previously undruggable since they haven’t had obvious active sites for small molecule binding. As well as their smaller size, this is a key differentiator to PROTACs. Approved MGDs have seen great commercial success, BMS’ Revlimid and pomalyst have 2023 projected sales of $6.5B and $847M, respectively.
Proprietary platform: Previous MGDs have been found by serendipity and a platform that enables their rationale design is needed. $GLUE has developed its QuEEN platform that uses a combination of AI and structural biology to identify targets that could particularly be amenable to MGDs and then identify chemical starting points for these targets. $GLUE has also developed bespoke assays to test and progress these molecules to candidate stage.
Evidence of success: The best evidence for the strength of a platform is progressing real molecules with differentiated profiles through hit-lead-candidate. Pipeline: MRT-2359 (GSPT1 degrader): is in PhI dose escalation for NSCLC, SCLC, DLBC and NE tumours. Demonstrated preclinical potency (other companies such as BMS have dropped GSPT1 degraders but Monte Rosa management affirms its asset is differentiated with respect to safety and pk). Monte Rosa also has a VAV1 and NEK7 degrader.
Feed early pipeline + investment in innovation: Hopefully the molecular glue approach can unlock novel targets to feed Roche’s early pipeline and build capabilities in AI.
Benefits to Monte Rosa:
Extends cash runway into Q3:25 funding the development of MRT-2359 and its pipeline of degraders
Enter into new therapeutic area of neuroscience - partnerships are a key way for a small biotech to apply their technologies to applicable areas without having to build up
Cellarity partners with Chan Zuckerberg to advance single-cell analysis
Partnerships such as this are great for attracting global talent from around the world to come together and progress the field of single-cell analysis, which is great for Cellarity’s mission.
Other highlighted deals:
Servier and Owkin partner to discover novel targets and indications
Thermofisher acquires Olink to expand proteomics capabilities
Eli Lilly continues it’s small ticket acquisition spree with Mablink
What we listened to
In case you missed it
A spotlight on the transformational science and innovation pioneered by UK life sciences companies
What we liked on Twitter
https://twitter.com/cziscience/status/1714717156708151775
https://twitter.com/CMichaelRose/status/1715026531637158343
Events
Field Trip
Did we miss anything? Would you like to contribute to Decoding Bio by writing a guest post? Drop us a note here or chat with us on Twitter: @ameekapadia @ketanyerneni @morgancheatham @pablolubroth @patricksmalone @zahrakhwaja











Perhaps the training images db is race vectored? Fastest solution is to change prompt.
Why do most of the people appear to be white in the AI generated picture?